Volume 11, Number 3—March 2005
Dispatch
Inquilinus limosus in Patients with Cystic Fibrosis, Germany
Abstract
We identified Inquilinus limosus, a recently described α-proteobacterium, in sputum of 2 patients with cystic fibrosis whose respiratory tracts were persistently colonized for >9 months. We present data on the epidemiology, antimicrobial susceptibility, and molecular characteristics of I. limosus.
Chronic microbial colonization of the respiratory tract, leading to exacerbations of pulmonary infection, is the major cause of disease and death in patients with cystic fibrosis (CF). Typical pathogens in respiratory secretions of patients with CF include Pseudomonas aeruginosa, Staphylococcus aureus, Haemophilus influenzae, and Burkholderia cepacia complex (1–3). Other gram-negative glucose nonfermenters, such as Achromobacter xylosoxidans, B. gladioli, Ralstonia pickettii, and Stenotrophomonas maltophilia are also occasionally recovered from respiratory samples from CF patients, but their pathogenic importance remains to be clarified (2–4).
Determining the clinical relevance of nonfermentative microbes is hampered by the difficulty in identifying these pathogens by conventional laboratory techniques. Recent studies that applied molecular approaches to identify unusual pathogens in patients with CF showed various infrequently encountered and novel species (4–7). In 1 of these studies, the gram-negative nonfermentative species Inquilinus limosus was newly described in respiratory secretions of 8 patients with CF in the United States (8). I. limosus belongs to the α-proteobacteria and is, thus, not closely related to B. cepacia complex or P. aeruginosa (8).
To detect unusual gram-negative microbes in respiratory samples, we screened all patients attending the CF clinic of the University Hospital, Ulm, Germany, from May 2002 to September 2004 (N = 85 patients). Respiratory samples (N = 459 samples) were plated with a 10-μL loop on sheep blood agar (Heipha, Heidelberg, Germany), MacConkey agar (Heipha), and B. cepacia complex selective agar (containing 100 mg/L ticarcillin and 300,000 IU/L polymyxin B, MAST Diagnostica, Reinfeld, Germany). All plates were incubated for 48 h at 36°C under ambient air, and the B. cepacia complex selective agar was incubated another 5 days at room temperature. I. limosus was recovered from sputum samples of 2 patients (0.9%).
Patient 1
Strain A was isolated from a 17-year-old male patient with CF with persistent colonization of the respiratory tract since childhood with S. aureus, P. aeruginosa, including the mucoid variant, and H. influenzae. In the initial sputum sample, apart from ≈106 CFU/mL of S. aureus, 105 CFU/mL of mucoid P. aeruginosa, and 103 CFU/mL of Candida albicans, 104 CFU/mL of a mucoid gram-negative rod was isolated from B. cepacia complex selective agar after 6 days of incubation. The isolate had positive oxidase and catalase reaction, but failed to grow on MacConkey agar. It was identified by using Api 20NE as Sphingomonas paucimobilis with questionable profile (Code 0427544). Final identification of I. limosus was achieved by sequencing the 16S rRNA gene with the primers 16Sfor and 16Srev (9) and a Dye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Warrington, United Kingdom) on a 310 Genetic Analyser (Abi Prism). The isolate showed 99.8% sequence homology to the 16S rDNA sequence of the I. limosus type strain (LMG 20952T) by using the BLAST algorithm.
During sequence analysis, we discovered a mistake in the I. limosus type strain 16S rDNA sequence deposited in GenBank (accession no. AY043374): the CGGGTC motif, repeated twice from base 956 to 967 in AY043374, is only found once in the type strain’s 16S rRNA gene, such as in the Inquilinus sp. strain AU1979 (accession no. AY043375 [8]) and in our isolates. We performed susceptibility testing by using Etest (VIVA Diagnostics, Solna, Sweden) on Mueller-Hinton agar, incubated at 36°C for 48 h. In addition, susceptibility testing against colistin was performed by disk diffusion with 10-μg disks (BD, Heidelberg, Germany) on Mueller-Hinton agar (McFarland 0.5, 48 h incubation). All results are shown in the Table. At the time of sputum sampling, the patient was clinically well, with normal values of C-reactive protein, leukocytes, and erythrocyte sedimentation rate. He regularly played soccer. A lung function test was not done. Two weeks after his visit, an elective 14-day course of antimicrobial therapy was initiated consisting of intravenous (IV) ceftazidime (3 g 3 times daily) and IV tobramycin (500 mg once daily) because of P. aeruginosa colonization.
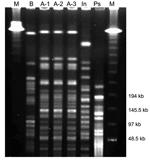
Five and a half months later, the patient was seen in the CF clinic again. He was still in very good clinical condition. Results of lung function tests conducted 6 weeks after his first visit and at his second visit were the following: vital capacity, 3.27l/3.34l (77%/74% of predicted vital capacity); and forced expiratory volume, 1 s 2.81l/2.89l (79%/77% of predicted forced expiratory volume). Sputum culture showed ≈106 CFU/mL of S. aureus, 104 CFU/mL of mucoid P. aeruginosa, 103 CFU/mL of Aspergillus fumigatus, and again 103 CFU/mL of I. limosus. We performed pulsed-field gel electrophoresis (PFGE) of the isolates with the CHEF DRIII equipment (BioRad, Munich,Germany) in 1% agarose at 14°C and a constant voltage of 200 V (10), with the restriction enzyme XbaI (11). Results showed that the strain (A-2) was identical to the former isolate of the same patient (A-1) (Figure). A 14-day course of oral ciprofloxacin (750 mg twice daily) was initiated because of P. aeruginosa colonization.
Four months later, the strain was still detected in his sputum. The isolate (A-3) grew in low quantities (103 CFU/mL) and was accompanied by 106 CFU/mL of S. aureus, 106 CFU/mL of mucoid P. aeruginosa, 103 CFU/mL of A. fumigatus, and 104 CFU/mL of C. albicans. PFGE showed identity with the former strains (Figure), and antimicrobial susceptibility was unchanged (Table). Lung function test showed a vital capacity of 3.81l (68% of predicted vital capacity) and a forced expiratory volume of 3.25l (71% of predicted forced expiratory volume), and the patient was in good health. More than 2 months later, the I. limosus strain was no longer cultured from his sputum, while P. aeruginosa (103 CFU/mL), S. aureus (106 CFU/mL), and C. albicans (103 CFU/mL) were still found.
Patient 2
Strain B was isolated from a sputum sample of a 14-year-old female patient with CF with respiratory colonization since childhood with P. aeruginosa, including the mucoid variant, and H. influenzae. The mucoid isolate of I. limosus grew in large quantities (≈105 CFU/mL) on B. cepacia complex selective agar after 5 days of incubation. In addition, ≈105 CFU/mL mucoid of P. aeruginosa, 103 CFU/mL of A. fumigatus and C. albicans were found in the sputum sample. Colonies were oxidase- and catalase-positive and failed to grow on MacConkey agar. Api 20NE showed Sphingomonas paucimobilis with questionable profile (Code 0424744), and identification was achieved by 16S rDNA sequencing, as described above. The isolate showed 99.3% sequence homology to the I. limosus type strain (LMG 20952T). PFGE showed a different band pattern, which suggests that this strain was different from strain A and, thus, excluded cross-infection between both patients (Figure).
The antimicrobial susceptibility of strain B was comparable to that of strain A, apart from higher MIC values for amikacin and gentamicin and lower values for ceftazidime (Table). Like the first patient, this patient was also in good health and active. At the time of sputum sampling, pulmonary function and laboratory tests were not done, but a pulmonary function test conducted 2 weeks before showed a vital capacity of 2.96l (74% of predicted vital capacity) and a forced expiratory volume 1 of 1.95l (77% of predicted forced expiratory volume 1), leukocyte count and erythrocyte sedimentation rate were normal, and C-reactive protein was slightly elevated to 5.8 mg/L. During the following 2 months, 6 sputum samples were investigated, but Inquilinus was not detected again, while P. aeruginosa was still present in a concentration of 105 CFU/mL. The patient moved to another CF clinic and was lost to follow-up.
To our knowledge, isolation of I. limosus from clinical samples has only been described in 1 study (8). The prevalence of this species in our CF clinic between May 2002 and September 2004 was 2.4%. The natural reservoir of I. limosus has yet to be discovered, but its relatedness to other nonfermentative rods suggests environmental sources. Inquilinus might be overlooked in clinical samples because of its rather slow growth and failure to grow on MacConkey agar. Recovery of Inquilinus can be improved by using selective media containing polymyxin B or colistin and ticarcillin, such as B. cepacia complex selective agar, and prolonged incubation at 36°C. The necessary duration of incubation has yet to be determined since our isolates grew better at 36°C than at room temperature. Identifying the species is difficult since it is not contained in the databases of commercial identification kits and its mucoid appearance may lead to confusion with mucoid P. aeruginosa strains. This species’ failure to grow on MacConkey agar, positive oxidase reaction, and typical antimicrobial susceptibility profile (see below) in respiratory samples of CF patients should arouse suspicion. Identification of I. limosus can be confirmed by 16S rRNA gene sequencing (8). I. limosus is able to persist in the respiratory tract of CF patients for several months. As with P. aeruginosa, abundant amounts of mucus with I. limosus infection may favor persistence and chronic infection. However, the pathogenic role of I. limosus in the patients described here is unclear. The stepwise deterioration of pulmonary function seen in patient 1 may also be attributed to irregular intervals of inhalation and of elective antimicrobial therapy. The patient had finished his education and had started his first employment.
I. limosus shows a distinct antimicrobial susceptibility profile with high MICs for cotrimoxazole, most aminoglycosides, ampicillin, cefotaxime, and piperacillin/tazobactam. Although ceftazidime and ciprofloxacin would be interpreted as susceptible applying the NCCLS interpretation criteria for P. aeruginosa (12), strain A persisted in the respiratory tract of the patient for several months after therapy with these substances. Inquilinus may be effectively protected from the action of antimicrobial agents by mucus production, and local host factors may also contribute to colonization and persistency. Further studies are necessary to evaluate the epidemiology and clinical importance of I. limosus as well as the therapeutic options in CF patients and in other patient groups. For instance, screening large CF patient groups by selective culture methods or molecular methods, like the use of specific fluorescence in situ hybridization probes or polymerase chain reaction assays, are desirable for assessing the epidemiology of the species. Longitudinal studies of infected patients are valuable in evaluating the clinical relevance and the factors influencing persistency of Inquilinus.
Acknowledgment
We thank Beate Wirths for excellent technical assistance and Angelika Möricke and Heike von Baum for performing PFGE.
References
- Klinger JD, Thomassen MJ. Occurrence and antimicrobial susceptibility of gram-negative nonfermentative bacilli in cystic fibrosis patients. Diagn Microbiol Infect Dis. 1985;3:149–58. DOIPubMedGoogle Scholar
- Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. DOIPubMedGoogle Scholar
- Miller MB, Gilligan PH. Laboratory aspects of management of chronic pulmonary infections in patients with cystic fibrosis. J Clin Microbiol. 2003;41:4009–15. DOIPubMedGoogle Scholar
- Coenye T, Vandamme P. LiPuma JJ. Infection by Ralstonia species in cystic fibrosis patients: identification of R. pickettii and R. mannitolilytica by polymerase chain reaction. Emerg Infect Dis. 2002;8:692–6.PubMedGoogle Scholar
- Coenye T, Liu L, Vandamme P. LiPuma JJ. Identification of Pandoraea species by 16S ribosomal DNA-based PCR assays. J Clin Microbiol. 2001;39:4452–5. DOIPubMedGoogle Scholar
- Coenye T, Vandamme P. LiPuma JJ. Ralstonia respiraculi sp. nov., isolated from the respiratory tract of cystic fibrosis patients. Int J Syst Evol Microbiol. 2003;53:1339–42. DOIPubMedGoogle Scholar
- Wallet F, Perez T, Armand S, Wallaert B, Courcol RJ. Pneumonia due to Bordetella bronchiseptica in a cystic fibrosis patient: 16S rRNA sequencing for diagnosis confirmation. J Clin Microbiol. 2002;40:2300–1. DOIPubMedGoogle Scholar
- Coenye T, Goris J, Spilker T, Vandamme P. LiPuma JJ. Characterization of unusual bacteria isolated from respiratory secretions of cystic fibrosis patients and description of Inquilinus limosus gen. nov., sp. nov. J Clin Microbiol. 2002;40:2062–9. DOIPubMedGoogle Scholar
- Hiraishi A. Direct automated sequencing of 16S rDNA amplified by polymerase chain reaction from bacterial cultures without DNA purification. Lett Appl Microbiol. 1992;15:210–3. DOIPubMedGoogle Scholar
- Pfaller MA, Holis RJ, Sader HS. PFGE of chromosomal DNA. In: Isenberg HD, editor. Clinical microbiology procedures handbook. Washington: American Society for Microbiology Press; p. 10.5.c.1-10.5.c.12.
- Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–9.PubMedGoogle Scholar
- NCCLS. Performance standards for antimicrobial susceptibility testing, MIC interpretation criteria for Pseudomonas aeruginosa, M100 S13. Wayne (PA): Institute of Clinical Laboratory Standards; 2003.
Figure
Table
Cite This ArticleTable of Contents – Volume 11, Number 3—March 2005
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Nele Wellinghausen, Department of Medical Microbiology and Hygiene, University of Ulm, Robert-Koch-Str. 8, D–89081, Ulm, Germany; fax: +49-(0)731-500 23473
Top