Volume 12, Number 12—December 2006
Dispatch
Mastomys natalensis and Lassa Fever, West Africa
Figure 2
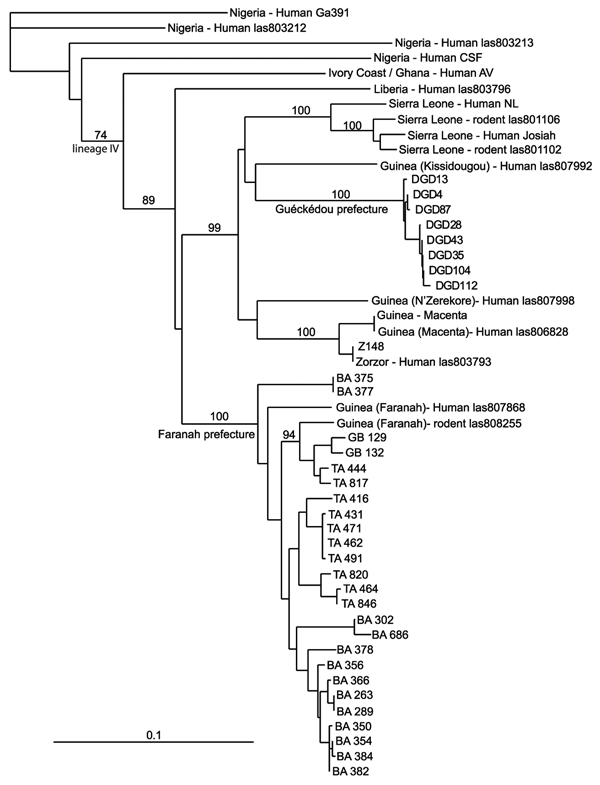
Figure 2. Phylogenetic relationships of Lassa virus strains based on a nucleoprotein gene fragment (631 bp) determined by using the neighbor-joining method. The numbers above branches are bootstrap values >50% (1,000 replicates). Scale bar indicates 10% divergence. Localities are indicated by the specimen label: DGD (Denguédou), BA (Bantou), GB (Gbetaya), and TA (Tanganya).
Page created: October 04, 2011
Page updated: October 04, 2011
Page reviewed: October 04, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.