Volume 12, Number 5—May 2006
Research
Novel Swine Influenza Virus Subtype H3N1, United States
Figure 2
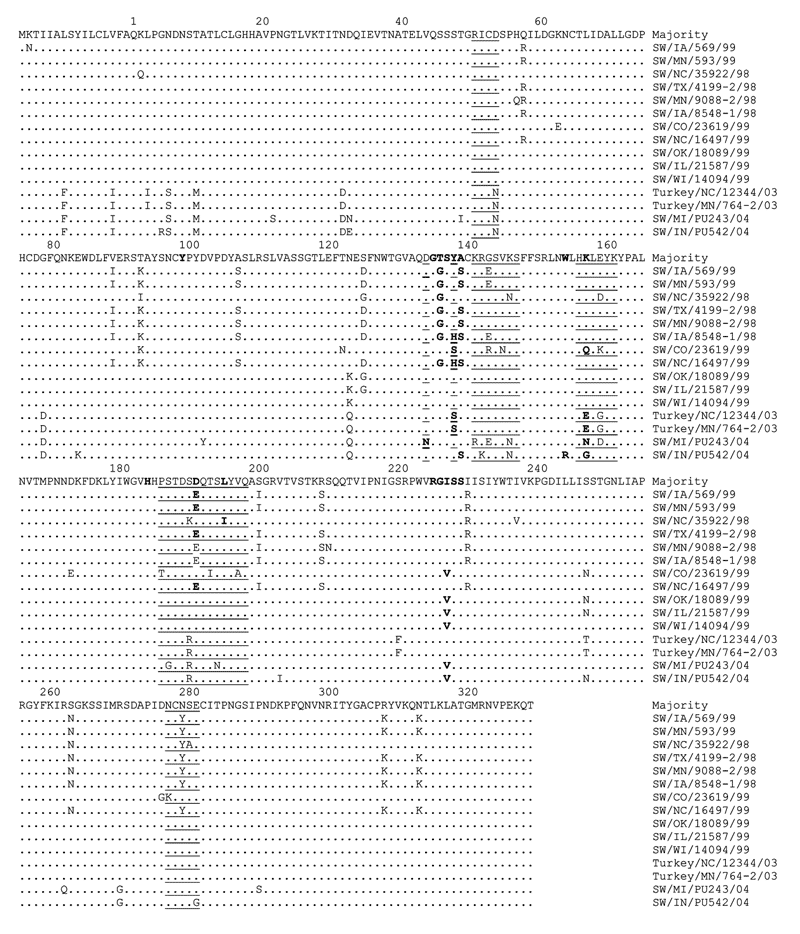
Figure 2. Alignment of deduced amino acid sequences within the hemagglutinin (HA) 1 region of HA genes of H3N2 swine influenza viruses (SIVs), H3N2 turkey isolates, and H3N1 SIVs. The amino acid sequence represents the consensus sequence, and the amino acid at position 1 is the first amino acid following the signal peptide (37). Dots represent amino acids similar to the consensus. Note that according to H3 structure (37), the residues representing the antigenic sites are underlined and the receptor binding sites are in boldface. The alignment shows that PU243 and PU542 may have emerged from the H3N2 turkey isolates. The residues within the receptor-binding site are relatively conserved.
References
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–79.PubMedGoogle Scholar
- Roger GN, Pritchette TJ, Lane JL, Paulson JC. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor infection: selection of receptor specific variants. Virology. 1983;131:394–408. DOIPubMedGoogle Scholar
- Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–8. DOIPubMedGoogle Scholar
- Guan Y, Shortridge KF, Krauss S, Li PH, Kawaoka Y, Webster RG. Emergence of avian H1N1 influenza viruses in pigs in China. J Virol. 1996;70:8041–6.PubMedGoogle Scholar
- Karasin AI, Anderson GA, Olsen CW. Isolation and characterization of H4N6 avian influenza viruses from pigs with pneumonia in Canada. J Virol. 2000;74:9322–7. DOIPubMedGoogle Scholar
- Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge KF, Potential for transmission of avian influenza viruses to pigs. J Gen Virol. 1994;75:2183–8. DOIPubMedGoogle Scholar
- Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol. 1998;72:7367–73.PubMedGoogle Scholar
- Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–73. DOIPubMedGoogle Scholar
- Matrosovich M, Tuzikov A, Bovin N, Gambaryan A, Klimov A, Castrucci MR, Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–12. DOIPubMedGoogle Scholar
- Vines A, Wells K, Matrosovich M, Castrucci MR, Ito T, Kawaoka Y. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host range restriction. J Virol. 1998;72:7626–31.PubMedGoogle Scholar
- Choi YK, Goyal SM, Joo HS. Prevalence of swine influenza virus subtypes on swine farms in the United States. Arch Virol. 2002;147:1209–20. DOIPubMedGoogle Scholar
- Shope RE. Swine Influenza. III. Filtration experiments and etiology. J Exp Med. 1931;54:373–80. DOIPubMedGoogle Scholar
- Karasin AI, Schutten MM, Cooper LA, Smith CB, Subbarao K, Anderson GA, Genetic characterization of H3N2 influenza viruses isolated from pigs in North America, 1977–1999: evidence for wholly human and reassortant virus genotypes. Virus Res. 2000;68:71–85. DOIPubMedGoogle Scholar
- Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999;73:8851–6.PubMedGoogle Scholar
- Richt JA, Lager KM, Clouser DF, Spackman E, Suarez DL, Yoon KJ. Real-time reverse transcription–polymerase chain reaction assays for the detection and differentiation of North American swine influenza viruses. J Vet Diagn Invest. 2004;16:367–73. DOIPubMedGoogle Scholar
- Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM, Webster RG. Evolution of swine H3N2 influenza viruses in the United States. J Virol. 2000;74:8243–51. DOIPubMedGoogle Scholar
- Karasin AI, Olsen CW, Anderson GA. Genetic characterization of an H1N2 influenza virus isolated from a pig in Indiana. J Clin Microbiol. 2000;38:2453–6.PubMedGoogle Scholar
- Karasin AI, Landgraf J, Swenson S, Erickson G, Goyal S, Woodruff M, Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J Clin Microbiol. 2002;40:1073–9. DOIPubMedGoogle Scholar
- Karasin AI, West K, Carman S, Olsen CW. Characterization of avian H3N3 and H1N1 influenza A viruses isolated from pigs in Canada. J Clin Microbiol. 2004;42:4349–54. DOIPubMedGoogle Scholar
- Choi YK, Goyal SM, Kang SW, Farnham MW, Joo HS. Detection and subtyping of swine influenza H1N1, H1N2 and H3N2 viruses in clinical samples using two-multiplex RT-PCR assays. J Virol Methods. 2002;102:53–9. DOIPubMedGoogle Scholar
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–89. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. DOIPubMedGoogle Scholar
- Choi YK, Lee JH, Erickson G, Goyal SM, Joo HS, Webster RG, H3N2 influenza virus transmission from swine to turkeys, United States. Emerg Infect Dis. 2004;10:2156–60.PubMedGoogle Scholar
- Richt JA, Lager KM, Janke BH, Woods RD, Webster RG, Webby RJ. Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J Clin Microbiol. 2003;41:3198–205. DOIPubMedGoogle Scholar
- Palmer DF, Coleman MT, Dowdle WR, Schild GC. Advanced laboratory techniques for influenza diagnosis. Immunology series no. 6. Washington: US Department of Health, Education, and Welfare. 1975. p. 51–2.
- Andreyev VG, Wesley RD, Mengeling WL, Vorwald AC, Lager KM. Genetic variation and phylogenetic relationships of 22 porcine reproductive and respiratory syndrome virus (PRRSV) field strains based on sequence analysis of open reading frame 5. Arch Virol. 1997;142:993–1001. DOIPubMedGoogle Scholar
- Stemke GW, Phan R, Young TF, Ross RF. Differentiation of Mycoplasma hyopneumoniae, M flocculare, and M hyorhinis on the basis of amplification of a 16S rRNA gene sequence. Am J Vet Res. 1994;55:81–4.PubMedGoogle Scholar
- Cooper L, Olsen C, Xu X. Molecular characterization of human influenza A viruses bearing swine-like hemagglutinin genes. Virus Evolution Workshop; 1999 Oct 21–24: Ardmore, Oklahoma.
- Kaverin NV, Rudneva IA, Ilyushina NA, Lipatov AS, Krauss S, Webster RG. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 escape mutants. J Virol. 2004;78:240–9. DOIPubMedGoogle Scholar
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. DOIPubMedGoogle Scholar
- Wright SM, Kawaoka Y, Sharp GB, Senne DA, Webster RG. Interspecies transmission and reassortment of influenza A viruses in pigs and turkeys in the United States. Am J Epidemiol. 1992;136:488–97.PubMedGoogle Scholar
- Andral B, Toquin D, Madec F, Aymard M, Gourreau JM, Kaiser C, Disease in turkeys associated with H1N1 influenza virus following an outbreak of the disease in pigs. Vet Rec. 1985;116:617–8. DOIPubMedGoogle Scholar
- Ficken MD, Guy JS, Gonder E. An outbreak of influenza (H1N1) in turkey breeder hens. Avian Dis. 1989;33:370–4. DOIPubMedGoogle Scholar
- Hinshaw VS, Webster RG, Bean WJ, Downie J, Senne DA. Swine influenza-like viruses in turkeys potential source of virus for humans? Science. 1983;220:206–8. DOIPubMedGoogle Scholar
- Suarez DL, Woolcock PR, Bermudez AJ, Senne DA. Isolation from turkey breeder hens of a reassortant H1N2 influenza virus with swine, human, and avian lineage genes. Avian Dis. 2002;46:111–21. DOIPubMedGoogle Scholar
- Tsai CP, Pan MJ. New H1N2 and H3N1 influenza viruses in Taiwanese pig herds. Vet Rec. 2003;153:408.PubMedGoogle Scholar
- Reid AH, Fanning TG, Janczewski TA, Taubenberger JK. Characterization of the 1918 “Spanish” influenza virus neuraminidase gene. Proc Natl Acad Sci U S A. 2000;97:6785–90. DOIPubMedGoogle Scholar
- Lindstrom S, Sugita S, Endo A, Ishida M, Huang P, Xi SH, Evolutionary characterization of recent human H3N2 influenza A isolates from Japan and China: novel changes in the receptor binding domain. Arch Virol. 1996;141:1349–55. DOIPubMedGoogle Scholar
- Baigent SJ, McCauley JW. Glycosylation of haemagglutinin and stalk-length of neuraminidase combine to regulate the growth of avian influenza viruses in tissue culture. Virus Res. 2001;79:177–85. DOIPubMedGoogle Scholar
- Kaverin NV, Gambaryan AS, Bovin NV, Rudneva IA, Shilov AA, Khodova OM, Postreassortment changes in influenza A virus hemagglutinin restoring HA-NA functional match. Virology. 1998;244:315–21. DOIPubMedGoogle Scholar
- Kobasa D, Wells K, Kawaoka Y. Amino acids responsible for the absolute sialidase activity of the influenza A virus neuraminidase: relationship to growth in the duck intestine. J Virol. 2001;75:11773–80. DOIPubMedGoogle Scholar
- Alexandra DJ. A review of avian influenza in different bird species. Vet Microbiol. 2000;74:3–13. DOIPubMedGoogle Scholar
1Current affiliation: Kasetsart University, Bangkok, Thailand