Volume 12, Number 8—August 2006
Dispatch
O'nyong-nyong Virus, Chad
Figure 2
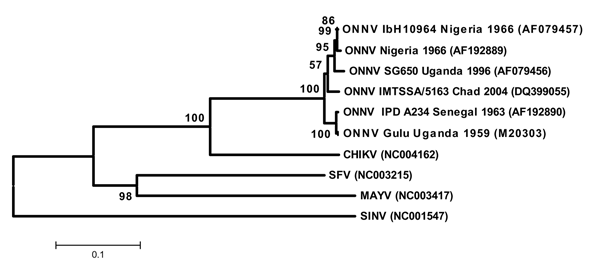
Figure 2. Phylogenetic tree of O'nyong-nyong virus (ONNV) based on partial E1 nucleotide sequence. Phylogram was constructed with MEGA 2 program (http://megasoftware.net/mega2.html) and tree drawing used the Juke-Cantor algorithm for genetic distance determination and the neighbor joining method. The percentage of successful bootstrap replicates (1,000 bootstrap replications, confidence probability >90%) is indicated at nodes. The length of the branches is proportional to the number of nucleotide changes (% of divergence). CHIKV (Chikungunya virus), SFV (Semliki Forest virus), MAYV (Mayaro virus), and SINV (Sindbis virus) sequences have been introduced for correct rooting of the tree.
Page created: December 09, 2011
Page updated: December 09, 2011
Page reviewed: December 09, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.