Volume 13, Number 10—October 2007
Research
Antigenic Diversity of Human Sapoviruses
Appendix Figure
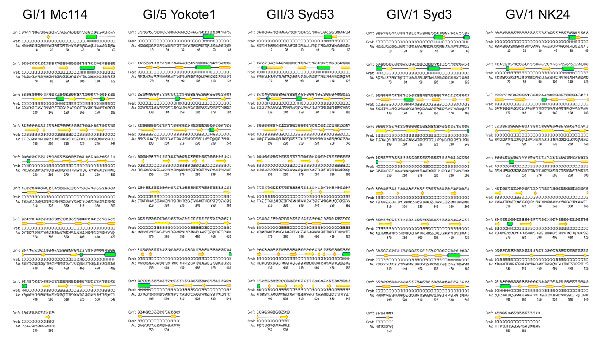
Appendix Figure. Schematic representations of complete predicted secondary structure of sapoviruses GI/1 Mc114, GI/5 Yokote1, GII/3 Syd53, GIV/1 Syd3, and GV/1 NK24 VP1. The first line shows level of confidence of prediction (Conf), where 10 represents high and 0 represents low confidence of prediction. The second line shows predicted secondary structure (Pred), where helix is indicated by a green cylinder, β strand by a yellow arrow, and coil by a line. The third line also shows predicted secondary structure (Pred), where helix is indicated by an H, β strand by an E, coil by a C. The fourth line shows amino acid (AA) sequence.
Page created: July 02, 2010
Page updated: July 02, 2010
Page reviewed: July 02, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.