Volume 13, Number 12—December 2007
Dispatch
Enhanced Subtyping Scheme for Salmonella Enteritidis
Abstract
To improve pulsed-field gel electrophoresis–based strain discrimination of 76 Salmonella Enteritidis strains, we evaluated 6 macro-restriction endonucleases, separately and in various combinations. One 3-enzyme subset, SfiI/PacI/NotI, was highly discriminatory. Five different indices, including the Simpson diversity index, supported this 3-enzyme combination for improved differentiation of S. Enteritidis.
Since 1987, Salmonella Enteritidis has been one of the most frequently isolated salmonellae associated with foodborne outbreaks (1). Illness from S. Enteritidis is linked to consumption of chickens, eggs, and foods that contain eggs (2). S. Enteritidis presents an interesting challenge from an epidemiologic perspective. Several reports documented a limited number of genotypes among ecologically diverse S. Enteritidis, reinforcing the notion that most S. Enteritidis strains are derived from a few endemic clones (3,4).
Pulsed-field gel electrophoresis (PFGE) is an integral subtyping tool used by several national public health networks (e.g., PulseNet, FoodNet, and VetNet) to differentiate outbreak strain clusters (5). The genetic homogeneity of S. Enteritidis, however, confounds many subtyping approaches, including PFGE (6,7). Conventional PFGE protocols lack discriminatory power to cull the subtle genotypic differences that distinguish S. Enteritidis strains. A more discriminatory scheme that incorporates combinations of potentially more informative enzymes may be attainable. We explored the discriminatory power of 6 enzymes, individually and in combination, to identify a more informative PFGE-based subtyping scheme for this important foodborne pathogen.
We examined 76 strains of S. Enteritidis and 74 strains of S. Typhimurium. Strains were isolated from poultry and poultry-related sources and were obtained from the Center for Veterinary Medicine and Center for Food Safety and Applied Nutrition of the US Food and Drug Administration and from the University of Georgia. After screening numerous restriction enzymes, the 6 selected were XbaI, BlnI, and SpeI, all used in PulseNet protocols (5); SfiI and PacI, previously used to improve resolution in PFGE studies involving Escherichia coli O157:H7 (8,9); and NotI, found to yield an optimal number of cut sites (10). The standard PulseNet PFGE protocol for non-typhoidal Salmonella was performed as described (11,12). Individual run conditions are listed in Table 1.
Five diversity indices were used to assess discriminatory potential among enzymes. First, unique PFGE patterns or pattern combinations, when analyzing >2 enzymes, were tallied. Second, the mean number of strains per polytomy (an unresolved strain cluster) was calculated as the number of polytomous strains divided by the number of polytomies in the tree. Third, the percentage of polytomous strains (of 76 S. Enteritidis strains) was calculated. Fourth, the node:strain ratio was calculated as the number of nodes (bifurcating tree forks) divided by 76 S. Enteritidis strains. A node:strain value closer to 1 indicated a more resolved tree. Finally, the Simpson diversity index was calculated as a numerical assessment of the relative discriminatory potential of each enzyme and enzyme combination (13).
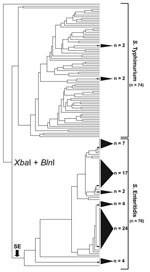
XbaI-BlnI patterns from 76 S. Enteritidis strains and 74 S. Typhimurium strains were analyzed simultaneously for a direct comparison of PFGE diversity. The resultant dendrogram yielded striking topologic differences between the 2 serovars (Figure 1). S. Typhimurium strains were almost entirely resolved; nearly every strain possessed its own branch on the dendrogram. In contrast, S. Enteritidis strain discrimination was sharply weaker, affirming extensive genetic homogeneity among strains. For example, 6 polytomies were evident in the S. Enteritidis portion of the tree, 5 of which comprised 4 or more strains and 1 of which comprised 24 strains. In total, 76% of S. Enteritidis strains occupied unresolved clusters with an average of ≈10 strains per cluster. Moreover, the S. Typhimurium dendrogram retained a nearly 1:1 ratio of nodes to strains, indicating that almost every strain retained a unique XbaI/BlnI pattern combination. S. Enteritidis, however, yielded a node:strain ratio of 1:3, indicating a relatively poorly bifurcated tree. Together, these observations highlighted the difficulty in differentiating S. Enteritidis with conventional PFGE approaches.
To develop a more discriminatory scheme for S. Enteritidis, we examined pattern diversity for 4 additional restriction endonucleases (SpeI, SfiI, PacI, and NotI). Diversity indices associated with each of the 6 enzymes are listed in Table 2. Many of the indices designated NotI as being effective for discriminating S. Enteritidis. Among the 6 enzymes, NotI yielded the highest number of unique patterns (n = 26), the fewest average number of strains per polytomy (5.2), the lowest percentage of strains captured by polytomies (82%), the highest node:strain ratio (0.47), and the highest Simpson diversity value (0.92). NotI was followed closely by PacI for most indices, which suggests that PacI was also useful for S. Enteritidis strain discrimination.
Previous studies that used PFGE noted the combining of restriction enzyme data into a single analysis as an approach for improving strain differentiation (8,9). In our study, a dendrogram of the combined 6-enzyme S. Enteritidis data was highly resolved and yielded 57 unique pattern combinations. The tree contained, on average, 3.6 strains per polytomy (n = 8), and only 38% of the strains in the 6-enzyme tree were associated with unresolved clusters (Table 2). The node:strain ratio was 0.78, and the Simpson index was 0.98, surpassing the accepted threshold (0.95) for a useful subtyping scheme (14).
Although highly effective for S. Enteritidis discrimination, 6-enzyme simultaneous analysis would increase the time and resources needed to complete investigations. Thus, a streamlined PFGE scheme with enhanced discrimination was desirable. First, 2-enzyme combinations were assessed to ascertain a minimal enzyme set for useful discrimination of S. Enteritidis. Combinations SfiI/NotI and PacI/NotI yielded optimal diversity measures, compared with the others (Table 2). However, because no 2-enzyme pair rivaled the discriminatory potential of the 6-enzyme analysis, 3-enzyme combinations were explored. A SfiI/PacI/NotI combination was found to be superior in all diversity categories (Table 2). It yielded 51 unique pattern combinations, an average of 3.3 strains per unresolved cluster, a node:strain ratio of 0.79, and a Simpson index of 0.98. The latter 3 measures matched or exceeded corresponding values in the 6-enzyme analysis, which indicates that most of the diversity in the 6-enzyme analysis was captured by this 3-enzyme combination.
The SfiI/PacI/NotI dendrogram showed a highly resolved tree topology and earmarked this 3-enzyme combination as being particularly effective for differentiating S. Enteritidis strains (Figure 2, panel A). Of 11 clusters, 9 comprised 3 or fewer strains, and only 47% of S. Enteritidis strains composed these 11 clusters, which, aside from the 6-enzyme analysis, represented the lowest portion of polytomous strains in the study.
The robust discrimination of S. Enteritidis achieved from combining SfiI, PacI, and NotI was attributed to polymorphic band classes that were lacking in other enzymes. As an example, the SfiI, PacI, and NotI PFGE patterns for 4 strains (S. Enteritidis 9, S. Enteritidis 12, 22704, and 22705) that were identical using XbaI and BlnI were examined for band variation. In contrast to XbaI, SfiI, PacI, and NotI, all showed some level of polymorphism among DNA fragments (Figure 2, panel B). Band differences for these 3 enzymes partitioned the 4 S. Enteritidis strains into disparate dendrogram positions.
On the basis of common geography and XbaI/BlnI pattern homogeneity, several clusters of S. Enteritidis strains appeared to exhibit clonal relatedness. However, the combined analysis of SfiI/PacI/NotI PFGE patterns was able to differentiate not only geographically disparate S. Enteritidis strains but also S. Enteritidis isolates from within a specific geographic locale (e.g., Georgia S. Enteritidis strains 415–421 and 434–436). Although several parameters, such as variable run conditions and times, likely preclude application to outbreak events in real time, the revised scheme presented here may be useful for retrospective epidemiologic investigations in which a more specific focus is often placed on a limited number of epidemiologically related isolates (15). Specifically, when S. Enteritidis strains are tightly linked geographically and temporally by clonal expansion, this 3-enzyme approach should provide an effective differentiation process.
Dr Zheng is a microbiologist and a faculty research associate at the University of Maryland, College Park, Maryland, USA. Her research interests include molecular characterization and pathogenesis of foodborne bacterial pathogens.
References
- Rodrigue DC, Tauxe RV, Rowe B. International increase in Salmonella Enteritidis: a new pandemic? Epidemiol Infect. 1990;105:21–7. DOIPubMedGoogle Scholar
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–12. DOIPubMedGoogle Scholar
- Olsen JE, Skov MN, Threlfall EJ, Brown DJ. Clonal lines of Salmonella enterica serotype Enteritidis documented by IS200-, ribo-, pulsed-field gel electrophoresis and RFLP typing. J Med Microbiol. 1994;40:15–22. DOIPubMedGoogle Scholar
- Saeed AM, Walk ST, Arshad M, Whittam TS. Clonal structure and variation in virulence of Salmonella Enteritidis isolated from mice, chickens, and humans. J AOAC Int. 2006;89:504–11.PubMedGoogle Scholar
- Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis. 2001;7:382–9.PubMedGoogle Scholar
- Thong KL, Ngeow YF, Altwegg M, Navaratnam P, Pang T. Molecular analysis of Salmonella Enteritidis by pulsed-field gel electrophoresis and ribotyping. J Clin Microbiol. 1995;33:1070–4.PubMedGoogle Scholar
- Landeras E, Gonzalez-Hevia MA, Alzugaray R, Mendoza MC. Epidemiological differentiation of pathogenic strains of Salmonella Enteritidis by ribotyping. J Clin Microbiol. 1996;34:2294–6.PubMedGoogle Scholar
- Davis MA, Hancock DD, Besser TE, Call DR. Evaluation of pulsed-field gel electrophoresis as a tool for determining the degree of genetic relatedness between strains of Escherichia coli O157:H7. J Clin Microbiol. 2003;41:1843–9. DOIPubMedGoogle Scholar
- Davis MA, Hancock DD, Besser TE, Rice DH, Hovde CJ, Digiacomo R, Correlation between geographic distance and genetic similarity in an international collection of bovine fecal Escherichia coli O157:H7 isolates. Epidemiol Infect. 2003;131:923–30. DOIPubMedGoogle Scholar
- Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 2005;33:D230–2. DOIPubMedGoogle Scholar
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis. Atlanta: The Centers; 1998 [cited 2007 Sep 20]. Available from http://www.cdc.gov/pulsenet/protocols.htm
- Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–6.PubMedGoogle Scholar
- Struelens MJ. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin Microbiol Infect. 1996;2:2–11.PubMedGoogle Scholar
- Schmid D, Luckner-Hornischer A, Holzhammer G, Rokita D, Federspeil M, Lassnig H, Lessons learned from a Salmonella Enteritidis phage type 4 outbreak in Austria, 2005. J Food Prot. 2007;70:35–9.PubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 13, Number 12—December 2007
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Eric W. Brown, Division of Microbiology, Center for Food Safety and Applied Nutrition, US Food and Drug Administration, Mailstop HFS-712, 5100 Paint Branch Pkwy, College Park, MD 20740, USA;
Top