Volume 13, Number 4—April 2007
Letter
Isolation of Schineria sp. from a Man
Figure
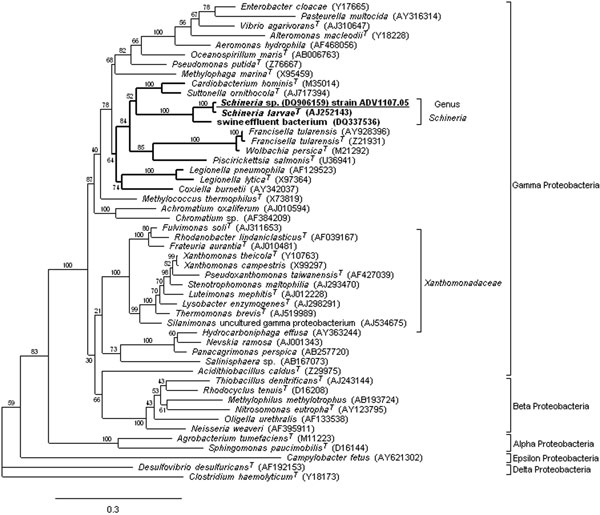
Figure. Maximum likelihood (ML) 16S rRNA gene phylogenetic tree showing the placement of the genus Schineria (boldface) and the isolate ADV1107.05 (underlined) in the phylum Proteobacteria. To reconstruct this tree, we used the strain ADV1107.05 sequence (DQ906159, 1441 bp) and 49 sequences selected from the GenBank database: 38 among the 15 orders of the Gamma Proteobacteria, 6 for the Beta Proteobacteria, 2 for the Alpha Proteobacteria, 1 for Delta Proteobacteria, 1 for Epsilon Proteobacteria and Clostridium haemolyticum (used as the outgroup organism). Accession nos. are in brackets. Alignment was performed with ClustalW 1.83 (4). ML phylogenetic analysis was performed by using PHYML v2.4.4 (5) with the general time-reversible plus gamma distribution plus invariable site (GTR + Γ + I) model found to be most appropriate according to Akaike information criteria. Bootstrap values given at the nodes are estimated with 100 replicates. The scale bar indicates 0.3 substitutions per nucleotide position. Strain ADV4155.05 sequence (DQ906158, 1414 bp) is not reported because it was identical to ADV1107.05. Trees were also obtained by distance methods (JC69, F84, and GTR models, and neighbor-joining), by parsimony, and by Bayesian inference. In all instances the genus Schineria branched out of the Xanthomonadaceae cluster.
References
- Toth E, Kovacs G, Schumann P, Kovacs AL, Steiner U, Halbritter A, Schineria larvae gen. nov., sp. nov., isolated from the 1st and 2nd larval stages of Wohlfahrtia magnifica (Diptera: Sarcophagidae). Int J Syst Evol Microbiol. 2001;51:401–7.PubMedGoogle Scholar
- Delhaes L, Bourel B, Scala L, Muanza B, Dutoit E, Wattel F, Case report: recovery of Calliphora vicina first-instar larvae from a human traumatic wound associated with a progressive necrotizing bacterial infection. Am J Trop Med Hyg. 2001;64:159–61.PubMedGoogle Scholar
- Teyssier C, Marchandin H, Jean-Pierre H, Diego I, Darbas H, Jeannot JL, Molecular and phenotypic features for identification of the opportunistic pathogens Ochrobactrum spp. J Med Microbiol. 2005;54:945–53. DOIPubMedGoogle Scholar
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. DOIPubMedGoogle Scholar
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. DOIPubMedGoogle Scholar
- Versalovic J, Koeuth T, Lupski JR. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–31. DOIPubMedGoogle Scholar
- Juteau P, Tremblay D, Villemur R, Bisaillon JG, Beaudet R. Analysis of the bacterial community inhabiting an aerobic thermophilic sequencing batch reactor (AT-SBR) treating swine waste. Appl Microbiol Biotechnol. 2004;66:115–22. DOIPubMedGoogle Scholar
- Saddler GS, Bradbury JF. Family I. Xanthomonadaceae fam. nov. In: Brenner DJ, Krieg NR, Staley JT, Garrity GM, editors. Bergey’s manual of systematic bacteriology. 2nd ed. Vol. 2. New York: Springer; 2005. p. 63.
- Mallon PW, Evans M, Hall M, Bailey R. Something moving in my head. Lancet. 1999;354:1260. DOIPubMedGoogle Scholar
- Nuesch R, Rahm G, Rudin W, Steffen I, Frei R, Rufli T, Clustering of bloodstream infections during maggot debridement therapy using contaminated larvae of Protophormia terraenovae. Infection. 2002;30:306–9. DOIPubMedGoogle Scholar