Volume 13, Number 6—June 2007
Dispatch
Norovirus Infection in Children with Acute Gastroenteritis, Madagascar, 2004–2005
Abstract
Of 237 children with acute gastroenteritis in Antananarivo, Madagascar, during May 2004–May 2005, 14 (≈6%) were infected with norovirus. Seasonality (November–December peak) was detected. Reverse transcription–PCR identified GII as the most common genogroup. GIs belonged to GI.1, GI.3, and GI.4. Noroviruses in Madagascar show extensive genetic diversity.
Gastroenteritis is a major public health issue worldwide. Noroviruses are now considered emerging pathogens (1) and are recognized as the leading cause of nonbacterial, acute gastroenteritis in humans (2). The 2 genera, Norovirus and Sapovirus, are members of the family Caliciviridae and have a positive-sense, single-stranded RNA genome ≈7.5 kb long. Because no readily available cell culture system exists, characterization and classification of noroviruses are based on reverse transcription (RT)–PCR, genomic sequencing, and phylogenetic analysis (2–5). According to the latest scheme for norovirus nomenclature, the 29 genetic norovirus clusters or genotypes are classified into 5 genogroups (GI–V) (6). GI and II infect humans, and GII.4 has been the most highly prevalent genotype worldwide during the past decade (2,5,7).
The saying “Madagascar is not an island, but an archipelago” captures an important aspect of the country, whose geography and history have combined to produce a society of considerable diversity and uniqueness (8). In recent years, many studies have investigated the role of human noroviruses in childhood diarrhea and found worldwide distribution (4,7,9,10). However, no studies have reported the prevalence and molecular epidemiology of noroviruses in Madagascar, which we report here.
From May 2004 to May 2005, a study of acute gastroenteritis in children <16 years of age was undertaken by the Institut Pasteur, Antananarivo, Madagascar. Children with a diagnosis of acute dehydrating watery diarrhea who were seen at the rehydration clinics and hospitals of Antananarivo were eligible for the study. Antananarivo, the capital city of Madagascar, has a population of ≈4 million. The study was approved by the Ethical Review Board of the Institut Pasteur, Antananarivo, Madagascar.
Fecal samples were collected from the children and stored at –80°C until analysis was undertaken at the University of Liverpool, UK. There, viral RNA was extracted from 150 μL of 10%–20% fecal suspensions in phosphate-buffered saline by using a guanidine and silica method (11). Sapoviruses were not sought in the initial screening. RT-PCR was performed in a 50-μL reaction mixture with primers that targeted the capsid N terminus/shell gene. For GI, forward primer G1SKF (5′-CTGCCCGAATTYGTAAATGA-3′) and reverse primer G1SKR (5′-CCAACCCARCCATTRTACA-3′) were used, yielding a PCR product of 330 bp. For GII, the primers used were forward primer G2SKF (5′-CNTGGGAGGGCGATCGCAA-3′) and reverse primers G2SKR (5′-CCRCCNGCATRHCCRTTRTACAT-3′) and G2ALSKR (5′-CCACCAGCATATGAATTGTACAT-3′), yielding a 344-bp product (12). The PCR was performed with an initial denaturation at 94°C, followed by 40 cycles of 60-sec denaturation at 94°C, 60-s primer annealing at 50°C, an extension for 2 min at 72°C, followed by a final extension stage of 15 min at 72°C. Amplification products were examined under ultraviolet light after electrophoresis through a 2% agarose gel with ethidium bromide staining. Products were extracted by using the QIAquick PCR purification kit (QIAGEN, Basingstoke, UK) and were sequenced by Macrogen Inc. (Seoul, South Korea).
Phylogenetic relationships were examined by aligning sequences with the ClustalW multiple alignment program (European Molecular Biology Laboratory, Heidelberg, Germany). A phylogenetic tree was constructed according to the neighbor-joining method by using ClustalX (version 1.83) and the alignment file obtained by analysis with ClustalW. Bootstrap values on a scale from 1 to 1,000 were also calculated. An unrooted phylogram of norovirus isolates from the present study and prototype strains was plotted in the PHYLIP format (http://evolution.genetics.washington.edu/phylip.html) output by using TreeView software, version 3.1 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Assignment of norovirus to genotype was made according to the scheme proposed by Zheng et al. (6). The nucleotide sequences of the Malagasy strains have been deposited at GenBank (accession nos. EF213624–EF213635, EF213638, and EF213640.) Norovirus-positive samples were screened for other viruses by negative-stain electron microsopy and by RT-PCR for rotavirus and astrovirus (13,14).
During this 12-month study, 258 children with acute gastroenteritis in Madagascar were screened for norovirus infection. Because 21 samples contained insufficient stool, 237 samples were analyzed (142 from boys and 95 from girls). Overall, 85% of children were <3 years of age, 77% were <2 years, 43% were <1 year, and 3% were newborns. The median age of the study population was 20 months (range 1 day to 16 years).
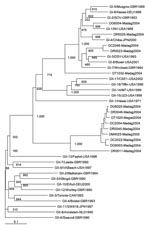
Fourteen noroviruses (5.9%) were detected in 237 children (Table 1). Noroviruses were found in all age groups. Infection rates did not differ between boys and girls (6.3% and 5.2%, respectively, p>0.1). No coinfections with other viruses (rotavirus, astrovirus, and adenovirus) were detected. Ten (71%) of the noroviruses detected were identified as belonging to genogroup GII; the remaining 4 (29%), to GI. Most GII noroviruses belonged to a potentially novel cluster (Figure 1). GI noroviruses were further classified into 3 genotypes: GI.1 (1 isolate), GI.4 (1 isolate), and GI.3 (2 isolates).
The median age of children with norovirus infection was 18 months (range 3–51 months). Most infections (≈86%) occurred in children <36 months of age. No infections were recorded in newborns or children >5 years of age. All infections with genogroup GI noroviruses were found in children <24 months of age (Table 2). Noroviruses were detected throughout the year; however, infections peaked during the wet season in Madagascar. November and December were the months of major norovirus prevalence (35.7% each month). GI noroviruses preceded GII infections by a few days (Figure 2).
To our knowledge, ours is the first study that has used molecular detection methods to investigate the role of noroviruses in pediatric gastroenteritis in Madagascar. We showed that infections with GI and GII noroviruses are relatively common. In a 1-year collection of stool samples, we detected noroviruses by RT-PCR in ≈6% of children with acute gastroenteritis in Antananarivo. This rate establishes norovirus as the second most commonly detected enteric virus in this population, behind rotavirus (38%) and followed by astrovirus (2.5%) (data not shown). These findings are consistent with those of studies elsewhere (7,10).
The median age of children with norovirus infection (18 months) was higher than previously reported (7) and higher than that of the rotavirus-infected group (median 10 months, range 1 day to 48 months) and that of the astrovirus-infected group (median 10 months, range 5–20 months) (data not shown). Noroviruses were detected throughout the year, but the number peaked in November and December. Such seasonality in a tropical country is not really expected, as year-round circulation has been previously documented (15).
Our findings confirm the finding of previous studies that GII is the predominant norovirus genogroup circulating in communities worldwide. Considerable genetic diversity was observed among the norovirus GII isolates, and some were identified as belonging to a potentially novel cluster. The closest reference strain to the potentially novel cluster was the recombinant Hu/NoV/GII.1/Hawaii/1971/US. In contrast, norovirus GI isolates were clustered with prototype strains; Hu/NoV/GI.3/DSV395/1990/SA (Desert Shield) was predominant (2 strains), followed by Hu/NoV/GI.1/Norwalk/1968/US (Norwalk) and Hu/NoV/GI.4/Chiba407/1987/JP (1 each).
This study has several limitations. First, it is a preliminary study. The sample size was small, and we examined samples collected over a 13-month period. Longer, longitudinal studies are required to address issues such as norovirus seasonality and temporal genetic variability. In addition, we restricted our analysis to specimens collected from patients at rehydration clinics and hospitals, so prevalence of norovirus infections in the general population may have been underestimated. Furthermore, the use of short conserved sequences, although successful for diagnosis of norovirus infection, should be used with caution for classification and phylogenetic analyses. Further analysis by full capsid sequencing might be required. Nevertheless, continued norovirus surveillance is needed to monitor the spread and persistence of the various genotypes infecting children in Madagascar.
Dr Papaventsis is an honorary clinical lecturer in medical microbiology and genitourinary medicine, Department of Host Defense and Infection, Faculty of Medicine, University of Liverpool, UK. His main research interests include the molecular epidemiology of enteric viruses.
Acknowledgment
We are grateful to P. Mauclere, for providing the opportunity to work with these samples. The study was funded internally by the University of Liverpool.
References
- Widdowson MA, Monroe SS, Glass RI. Are noroviruses emerging? Emerg Infect Dis. 2005;11:735–7.PubMedGoogle Scholar
- Radford AD, Gaskell RM, Hart CA. Human norovirus infection and the lessons from animal caliciviruses. Curr Opin Infect Dis. 2004;17:471–8. DOIPubMedGoogle Scholar
- Ando T, Noel JS, Fankhauser RL. Genetic classification of “Norwalk-like viruses.”. J Infect Dis. 2000;181(Suppl2):S336–48. DOIPubMedGoogle Scholar
- Zintz C, Bok K, Parada E, Barnes-Eley M, Berke T, Staat MA, Prevalence and genetic characterization of caliciviruses among children hospitalized for acute gastroenteritis in the United States. Infect Genet Evol. 2005;5:281–90. DOIPubMedGoogle Scholar
- Kageyama T, Shinohara M, Uchida K, Fukushi S, Hoshino FB, Kojima S, Coexistence of multiple genotypes, including newly identified genotypes, in outbreaks of gastroenteritis due to norovirus in Japan. J Clin Microbiol. 2004;42:2988–95. DOIPubMedGoogle Scholar
- Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006;346:312–23. DOIPubMedGoogle Scholar
- Dove W, Cunliffe NA, Gondwe JS, Broadhead RL, Molyneux ME, Nakagomi O, Detection and characterization of human caliciviruses in hospitalized children with acute gastroenteritis in Blantyre, Malawi. J Med Virol. 2005;77:522–7. DOIPubMedGoogle Scholar
- Covell M. Geographical and historical background. In: Madagascar: politics, economics and society. London: Pinter Pub Ltd; 1987. p. 8.
- Hansman GS, Katayama K, Maneekarn N, Peerakome S, Khamrin P, Tonusin S, Genetic diversity of norovirus and sapovirus in hospitalized infants with sporadic cases of acute gastroenteritis in Chiang Mai, Thailand. J Clin Microbiol. 2004;42:1305–7. DOIPubMedGoogle Scholar
- Bereciartu A, Bok K, Gomez J. Identification of viral agents causing gastroenteritis among children in Buenos Aires, Argentina. J Clin Virol. 2002;25:197–203. DOIPubMedGoogle Scholar
- Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-Van Dillen PM, Van Der Noordaa J. Rapid and simple method for purification of nucleic acids. J Clin Microbiol. 1990;28:495–503.PubMedGoogle Scholar
- Kojima S, Kageyama T, Fukushi S, Hoshimo FB, Shinohara M, Uchida K, Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. 2002;100:107–14. DOIPubMedGoogle Scholar
- Cunliffe NA, Dove W, Bunn JE, Ben Ramadan M, Nyangao JW, Riveron RL, Expanding global distribution of rotavirus serotype G9: detection in Libya, Kenya, and Cuba. Emerg Infect Dis. 2001;7:890–2. DOIPubMedGoogle Scholar
- Cunliffe NA, Dove W, Gondwe JS, Thindwa BD, Greensill J, Holmes JL, Detection and characterization of human astroviruses in children with acute gastroenteritis in Malawi. J Med Virol. 2002;67:563–6. DOIPubMedGoogle Scholar
- O’Ryan ML, Mamani N, Gaggero A, Avendano LF, Prieto S, Pena A, Human caliciviruses are a significant pathogen of acute sporadic diarrhea in children of Santiago, Chile. J Infect Dis. 2000;182:1519–22. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 13, Number 6—June 2007
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
C. Anthony Hart, Department of Medical Microbiology and Genito-Urinary Medicine, University of Liverpool, Duncan Building, Daulby St, Liverpool L69 3GA, UK;
Top