Volume 14, Number 3—March 2008
Research
High Rate of Mobilization for blaCTX-Ms
Figure 1
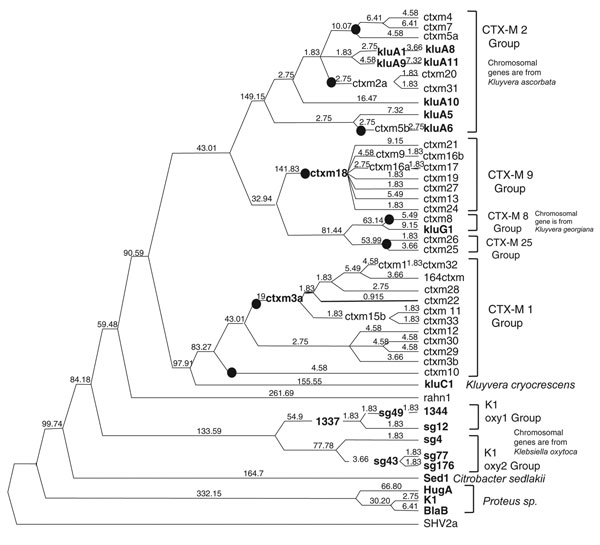
Figure 1. Phylogenetic analysis of blaCTX-Ms. This tree was calculated by Bayesian inference. Number of mutations occurring along each branch are given along the length of the branch. Black dots represent mobilizations. Boldface indicates chromosomal genes. CTX-M-14 and 3a exist as both unmobilized chromosomal genes and plasmid-borne CTX-M alleles.
Page created: July 07, 2010
Page updated: February 26, 2015
Page reviewed: February 26, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.