Volume 15, Number 10—October 2009
Dispatch
Diversity and Origin of Dengue Virus Serotypes 1, 2, and 3, Bhutan
Figure 2
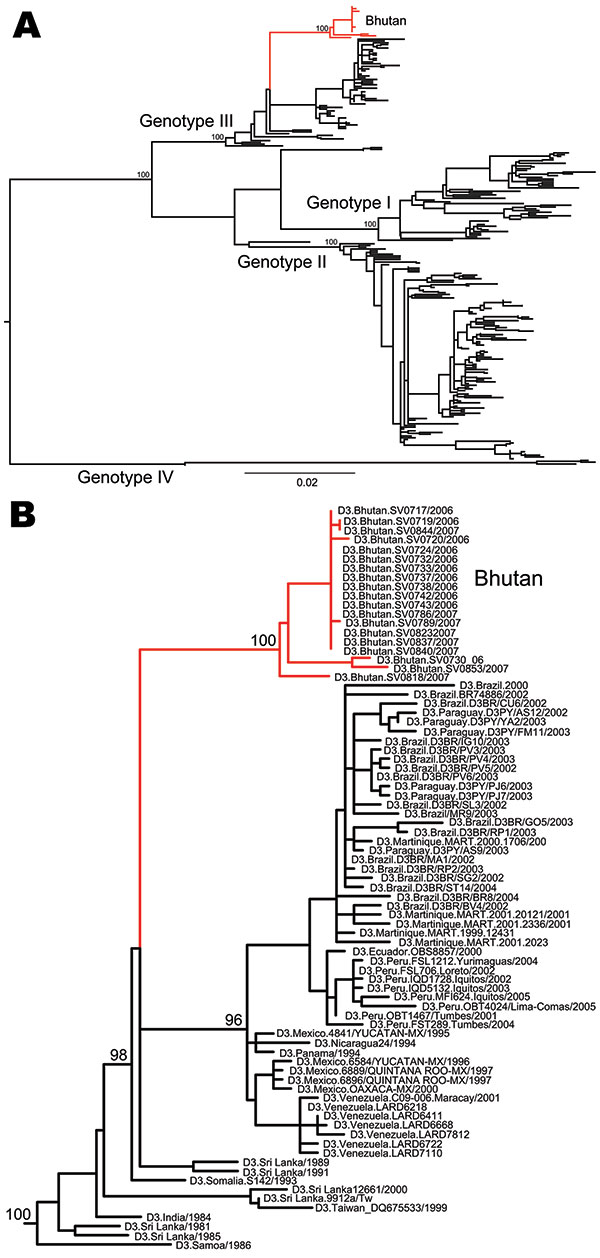
Figure 2. A) Maximum-likelihood phylogenetic tree of 264 complete envelope gene sequences of dengue virus serotype 3 (DENV-3). The different genotypes of DENV-3 and the isolates from Bhutan (red) are indicated. Scale bar indicates number of substitutions per site. B) Magnification of the part of the phylogeny where the Bhutan sequences (red) fall. The tree is midpoint rooted for clarity only, and all horizontal branch lengths are drawn to a scale of nucleotide substitutions per site. Bootstrap support values are shown for key nodes only.
Page created: December 08, 2010
Page updated: December 08, 2010
Page reviewed: December 08, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.