Volume 15, Number 11—November 2009
Letter
Dengue Virus Type 3 Infection in Traveler Returning from West Africa
Figure
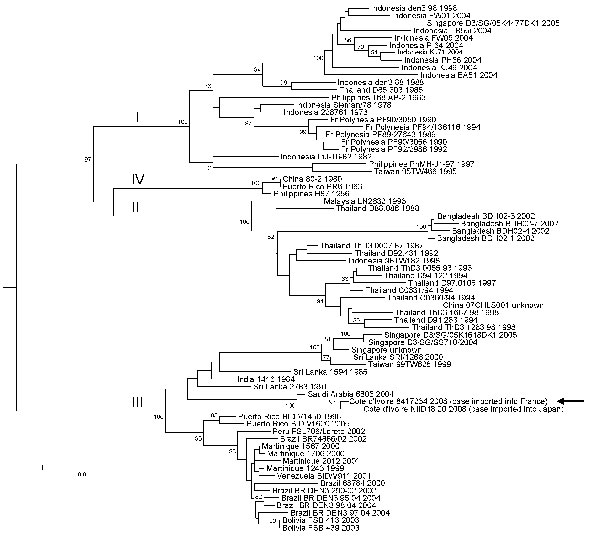
Figure. Phylogenetic analysis of selected dengue virus type 3 (DENV-3) sequences. The main genotypes are indicated using roman numerals at the node of the lineage. Sequence identification is as follows: country of origin, strain name, year of isolation/detection. The sequence determined in our study is underlined and designated by an arrow. Phylogenetic studies were conducted by using MEGA version 2.1 (5). Genetic distances were calculated with the Kimura 2-parameter method at the nucleotide level. Phylogenetic trees were constructed using the neighbor-joining method. The robustness of the nodes was tested by 500 bootstrap replications. The tree was rooted with DENV-1, DENV-2, and DENV-4 sequences. Scale bar indicates nucleotide substitutions per site.
References
- Schwartz E, Weld LH, Wilder-Smith A, von Sonnenburg F, Keystone JS, Kain KC, GeoSentinel Surveillance Network. Seasonality, annual trends, and characteristics of dengue among ill returned travelers, 1997–2006. Emerg Infect Dis. 2008;14:1081–8. DOIPubMedGoogle Scholar
- Fulhorst CF, Monroe MC, Salas RA, Duno G, Utrera A, Ksiazek TG, Isolation, characterization and geographic distribution of Cano Delgadito virus, a newly discovered South American hantavirus (family Bunyaviridae). Virus Res. 1997;51:159–71. DOIPubMedGoogle Scholar
- Moureau G, Temmam S, Gonzalez JP, Charrel RN, Grard G, de Lamballerie X. A real-time RT-PCR method for the universal detection and identification of flaviviruses. Vector Borne Zoonotic Dis. 2007;7:467–77. DOIPubMedGoogle Scholar
- Leparc-Goffart I, Baragatti M, Temman S, Tuiskunen A, Moureau G, Charrel R, Development and validation of real time one-step reverse transcription-PCR for the detection and typing of dengue viruses. J Clin Virol. 2009;45:61–6. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: Molecular Evolutionary Genetics Analysis software. Tempe (AZ): Arizona State University; 2001.
- Dengue in Africa. emergence of DENV-3, Côte d’Ivoire, 2008. Wkly Epidemiol Rec. 2009;84:85–8.PubMedGoogle Scholar
- Durand JP, Vallée L, de Pina JJ, Tolou H. Isolation of a dengue type 1 virus from a soldier in West Africa (Côte d’Ivoire). Emerg Infect Dis. 2000;6:83–4. DOIPubMedGoogle Scholar
- Leroy EM, Nkogue D, Ollomo B, Nze-Nkogue C, Becquart P, Grard G, Concurrent chikungunya and dengue virus infections during simultaneous outbreaks, Gabon, 2007. Emerg Infect Dis. 2009;15:591–3. DOIPubMedGoogle Scholar
- Messer WB, Gubler DJ, Harris E, Sivananthan K, de Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003;9:800–9.PubMedGoogle Scholar
- Freedman DO, Weld LH, Kozarsky PE, Fisk T, Robins R, von Sonnenburg F, Spectrum of disease and relation to place of exposure among ill returned travelers. N Engl J Med. 2006;354:119–30. DOIPubMedGoogle Scholar