Volume 15, Number 2—February 2009
Research
Simian T-Lymphotropic Virus Diversity among Nonhuman Primates, Cameroon
Figure 2
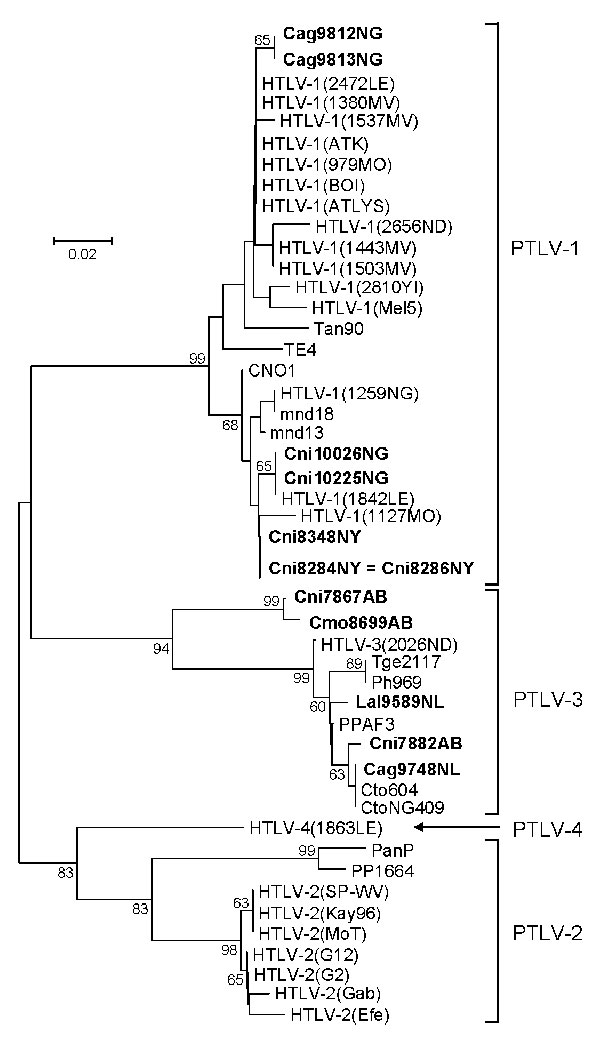
Figure 2. Primate T-lymphotropic virus (PTLV) phylogeny inferred by using 161-bp tax sequences. New sequences from nonhuman primates (NHPs) from Cameroon in this study are in boldface. Support for the branching order was determined by 1,000 bootstrap replicates; only values >60% are shown. Branch lengths are proportional to the evolutionary distance (scale bar) between the taxa. Nonhuman primates are coded as follows: the first letter of the genus is followed by the first 2 letters of the species name: Cmo, Cercopithecus mona; Cni, Cercopithecus nictitans; Cto, Cercocebus torquatus; Ppa, Papio papio; Ph, Papio hamadryas; Tge, Theropithecus gelada. Cag, Cercocebus agilis; Lal, Lophocebus albigena; Mnd and msp, Mandrillus sphinx; PanP and PP, Pan paniscus; Ptr, Pan troglodytes; Ggo, Gorilla gorilla; Tan, tantalus monkey; Cag, Cercocebus agilis; Mar, Macaca arctoides; Pha, Papio hamadryas; Pan, Papio anubis; Bab, baboon; HYB, hybrid baboon (Pha X Pan); Cae, Chlorocebus aethiops (AGM, African green monkey); Cpo, Cercopithecus pogonias; Cmi, Cercopithecus mitis; Cce, Cercopithecus cephus; Ang, Allenopithecus nigroviridis; Wrc, Western red colobus. The last 2 letters in the code indicate the study site: AB, Abat; MV, Mvangan; NY, Nyabissan; NL, Ngoila; MN, Manyemen; BA, Bangourain; MA, Massangam; YI, Yingui; ND, Ndikinimeki; NG, Ngovayan; SA, Sobia; LE, Lomie; MO, Mouloundou.
1Current affiliation: Stanford University, Stanford, California, USA.
2Current affiliation: University of Pittsburgh Graduate School of Public Health, Pittsburgh, Pennsylvania, USA.