Volume 15, Number 8—August 2009
Dispatch
Epidemiologic Study of Vibrio vulnificus Infections by Using Variable Number Tandem Repeats
Figure 1
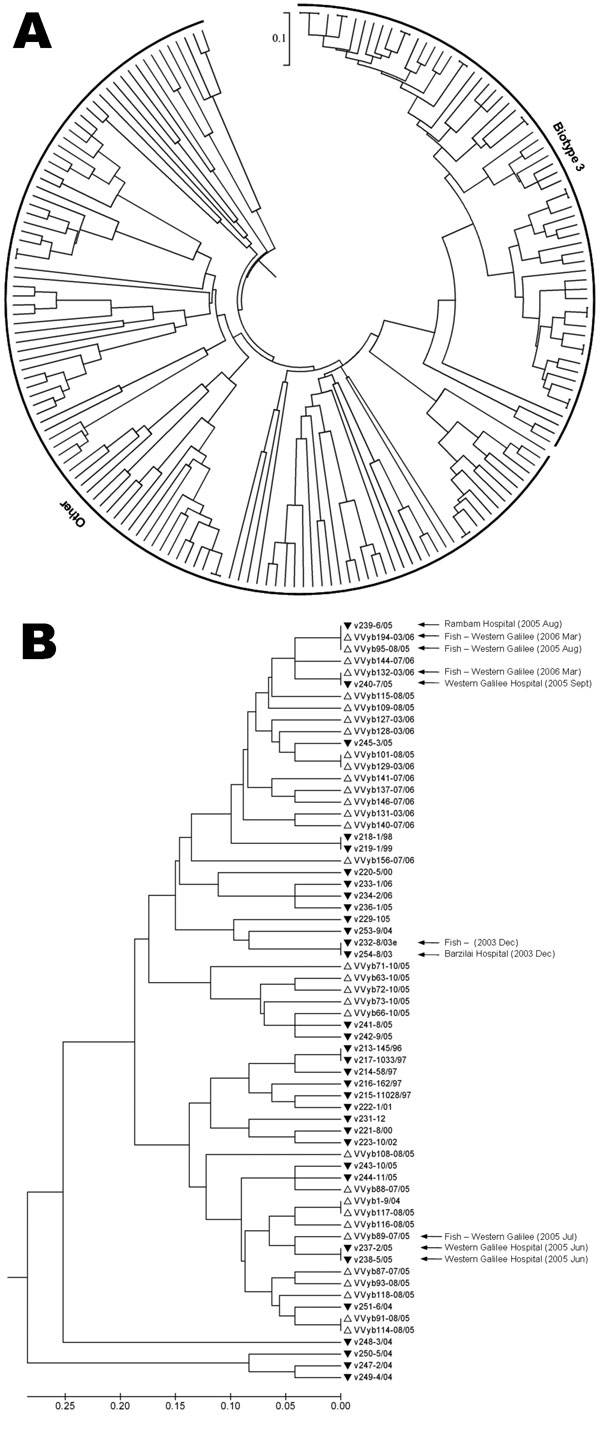
Figure 1. A) Genetic relationships based on simple-sequence repeat (SSR) variation data among 183 Vibrio vulnificus isolates including 135 new environmental, 22 new clinical, and 26 previously studied isolates. B) A subtree enlargement of panel A displaying a set of 65 V. vulnificus biotype 3 isolates. Similar clinical and environmental isolates, showing an epidemiologic connection, are indicated by arrows. The genetic-distance matrix was generated based on 212 polymorphic points (the sum of alleles across 12 SSR loci). Genetic relationships are based on unweighted pair group method with arithmetic mean cluster analysis of SSR variation using MEGA4 software (11). Scale bar represents genetic distance.
References
- Farmer JJ. Vibrio. In: Patrick RM, Ellen JB, James HJ, Michael AP, Robert HY, editors. Manual of clinical microbiology. Washington: American Society for Microbiology Press; 2003. p. 706–16.
- Oliver JD. Vibrio vulnificus. In: Doyle MP, editor. Food borne bacterial pathogens. New York: Marcel Dekker Inc.; 1989. p. 569–600.
- Bisharat N, Agmon V, Finkelstein R, Raz R, Ben Dror G, Lerner L, Clinical, epidemiological, and microbiological features of Vibrio vulnificus biogroup 3 causing outbreaks of wound infection and bacteraemia in Israel. Lancet. 1999;354:1421–4. DOIPubMedGoogle Scholar
- Bisharat N, Cohen DI, Harding RM, Falush D, Crook DW, Peto T, Hybrid Vibrio vulnificus. Emerg Infect Dis. 2005;11:30–5.PubMedGoogle Scholar
- Bisharat N, Amaro C, Fouz B, Llorens A, Cohen DI. Serological and molecular characteristics of Vibrio vulnificus biotype 3: evidence for high clonality. Microbiology. 2007;153:847–56. DOIPubMedGoogle Scholar
- Chatzidaki-Livanis M, Hubbard MA, Gordon K, Harwood VJ, Wright AC. Genetic distinctions among clinical and environmental strains of Vibrio vulnificus. Appl Environ Microbiol. 2006;72:6136–41. DOIPubMedGoogle Scholar
- Broza YY, Danin-Poleg Y, Lerner L, Broza M, Kashi Y. Vibrio vulnificus typing based on simple sequence repeats: insights into the biotype 3 group. J Clin Microbiol. 2007;45:2951–9. DOIPubMedGoogle Scholar
- Zaidenstein R, Sadik C, Lerner L, Valinsky L, Kopelowitz J, Yishai R, Clinical characteristics and molecular subtyping of Vibrio vulnificus illnesses, Israel. Emerg Infect Dis. 2008;14:1875–82. DOIPubMedGoogle Scholar
- Danin-Poleg Y, Cohen LA, Gancz H, Broza YY, Goldshmidt H, Malul E, Vibrio cholerae strain typing and phylogeny study based on simple sequence repeats. J Clin Microbiol. 2007;45:736–46. DOIPubMedGoogle Scholar
- Torpdahl M, Sorensen G, Lindstedt BA, Nielsen EM. Tandem repeat analysis for surveillance of human Salmonella typhimurium infections. Emerg Infect Dis. 2007;13:388–95. DOIPubMedGoogle Scholar
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186:1518–30. DOIPubMedGoogle Scholar
- Rosche TM, Yano Y, Oliver JD. A rapid and simple PCR analysis indicates there are two subgroups of Vibrio vulnificus which correlate with clinical or environmental isolation. Microbiol Immunol. 2005;49:381–9.PubMedGoogle Scholar
- Oliver JD. The viable but nonculturable state in bacteria. J Microbiol. 2005;43:93–100.PubMedGoogle Scholar
- Randa MA, Polz MF, Lim E. Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl Environ Microbiol. 2004;70:5469–76. DOIPubMedGoogle Scholar
Page created: November 01, 2010
Page updated: November 01, 2010
Page reviewed: November 01, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.