Volume 16, Number 10—October 2010
Dispatch
Hepatitis E Virus Genotype Diversity in Eastern China
Abstract
We studied 47 hepatitis E virus (HEV) isolates from hospitalized patients in Nanjing and Taizhou, eastern China. Genotypes 1, 3, and 4 were prevalent; genotype 3 and subgenotype 4b showed a close relationship with the swine strains in eastern China, thus indicating that HEV genotype 3 had infected humans in China.
Hepatitis E virus (HEV), genus Hepevirus, is a nonenveloped virus with a positive-stranded RNA genome of ≈7.2 kb (1). Researchers have hypothesized that zoonotic infection is involved in HEV transmission (2,3). HEV isolates have been divided into 4 distinct genotypes, and further classification of the 4 genotypes into 24 subtypes has been proposed (4). Genotypes 1 and 2 have been identified exclusively in humans, and genotypes 3 and 4 have been found in humans and several animal species. Genotypes 1 and 2 have been isolated in Asia, Africa, and North America; genotype 4 has been identified only in Asia; and genotype 3 has been found in almost every country. Although genotype 3 was found to be prevalent in swine populations in China in 2007 (5), this viral genotype has not previously been reported in humans in this country. Our study aimed to characterize the genotype diversity of the strains and to determine the full-length sequence of the most prevalent genotype in eastern China.
We studied 47 HEV isolates from patients (15 women and 32 men, 19–73 years of age) admitted to hospitals in Nanjing (26 patients) and Taizhou (21 patients) in eastern China and who were serologically confirmed to have HEV infection by commercial ELISAs (Wan Tai Pharmaceutical, Beijing, People’s Republic of China). HEV RNA was detected by reverse transcription–PCR as described (6). A serum sample negative for HEV was included as a control for testing sample contamination.
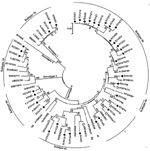
Sequence analysis based on the PCR-amplified products (primer sequences were removed) indicated that among the 47 HEV strains, 31 belonged to genotype 4, 13 belonged to genotype 1, and 3 belonged to genotype 3, which indicated that genotype 4 is the main type prevalent in humans in eastern China. The 13 genotype 1 isolates shared >98% sequence homology and showed 3 distinct nucleotide sequences. Figure 1 shows the phylogenetic tree constructed for the 37 distinct sequences in this study and their closest matching sequences in GenBank. To further classify the subgenotype of these strains, we included in the phylogenetic analysis some well-characterized subtype strains (4).
Results indicated that the 31 genotype 4 isolates found in this study were divided into 5 different subtypes. Seventeen of the genotype 4 isolates were subtyped as 4b, according to the reference sequences (GenBank accession nos. AJ344188 and AB116161), and they shared 93.2%–99.7% sequence homology among themselves. Strains ECh250 shared 100% sequence homology with FJ461766 and could be classified into subtype 4e, according to the reference sequence (AF234501). Five of the isolates, which shared 89.4%–91.2% sequence identities, belonged to subtype 4d, defined by reference strain AB082559. Four of the strains shared 92.4%–95.7% sequence homology and could be defined as subtype 4a, according to the reference stsrain (AJ344171).
Notably, the other 4 genotype 4 isolates could not be subtyped on the basis of the reference strains described by Lu et al. (4) but closely clustered with each other, forming a new subtype (Figure 1). The 3 genotype 1 isolates identified in this study shared >98% sequence homology and could be defined as subtype 1b, a virus subtype mainly prevalent in China (4). The 3 genotype 3 isolates in this study shared 97.7%–98.6% sequence identities and were closely related (96.4%–97.9% sequence identities) to a swine isolate (FJ527832), found in swine in the Shanghai area (7).
Genotype 4 HEV has been found to be the dominant cause of hepatitis E in China and has been involved in zoonotic transmission in eastern and southern China (3,8). When a BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) search in GenBank was performed, we found that some genotype 4 isolates obtained in this study shared the highest sequence homology with previous swine strains (Figure 1). For subtypes 4a, 4c, and 4e and the new subtype groups, the swine strains that showed the closest relationship with our studied strains had been isolated from swine in other regions of China; this finding suggests that these isolates might not be indigenous to eastern China (Figure 1). Fifteen of the genotype 4 isolates in the 4b group had >96.3% sequence identity and closely clustered with a swine strain (EU375332) that had been isolated from swine in eastern China (9); strain ECh118 had 99.3% sequence homology to this swine strain (Figure 1).
Because the serum specimen from which ECh118 was identified was insufficient for complete genome amplification, EChZ20 was selected for complete genome sequencing using the primers described previously (10). The complete genome comprises 7,228 nt, excluding the 3′ poly(A) tail. The open reading frame 1 (ORF1) begins at nt 26 and ends at nt 5140 (5,115 nt); ORF2 (nt 5137–7161) comprises 2,022 nt and encodes 674 aa; ORF3 (nt 5165–5509) comprises 345 nt and encodes 114 aa.
The phylogenetic tree obtained for the complete genome of EChZ20 and other 16 representative genotype 4 isolates indicated that EChZ20 closely clustered with DQ450072, EF570133, and AB369690 and shared 90.7%, 91.7%, and 89.1% sequence identities with them, respectively (Figure 2, panel A). DQ450072 and EF570133 were isolated from swine in eastern China (10), and AB369690 was isolated from a patient from Japan who had traveled to Shanghai, China, before the onset of acute hepatitis E, according to information in GenBank. These results suggested that subtype 4b virus isolates were involved in cross-species transmission from swine to humans in eastern China.
The genotype 3 isolates found in many countries, whether from swine or humans, have been reported to show a strong genetic relationship (11–13). Since 2007, genotype 3 has been found in swine groups in several areas in China (14); however, no reports have indicated that genotype 3 HEV infects humans in this country. In our study, genotype 3 was detected in humans in eastern China, and the strains found in humans showed high sequence homology (96.4%–97.9%) to the virus strains prevalent in swine in China. We intend to amplify the complete genome of this virus strain; however, because the quantity of serum in this study was limited, only a 1,681-nt partial ORF2 sequence of EChN22 was obtained.
Phylogenetic analysis, based on the 1681-nt sequence of EChN22 and other genotype 3 strains available in GenBank, confirmed that EChN22 belonged to genotype 3b and that it clustered closely with a swine isolate from China (FJ527832) and shared 97.2% nt and 99.6% aa sequence homology with it (Figure 2, panel B). These results suggested that the genotype 3 virus strain prevalent in humans in eastern China might come from swine in this area. The sequences were deposited in GenBank under accession nos. HM439249–HM439285
Our study indicated that 3 genotypes of HEV (1, 3, and 4) are prevalent in humans in eastern China. The 31 genotype 4 strains could be further divided into 5 different subtypes, including a new subtype that, to our knowledge, has not been subtyped in previous studies. The complete genome of 1 representative strain of the most prevalent subtype (genotype 4b) was sequenced, and it showed a close relationship with swine strains prevalent in swine in eastern China.
Three genotype 3 HEV strains were identified, and one of them was selected and sequenced to obtain a longer sequence (1,681 nt). Phylogenetic analysis based on the 1,681-nt partial ORF2 sequence indicated the genotype 3 strain showed relatively high sequence homology (97.2%) to virus strains recently isolated from swine in eastern China. This finding suggests that the genotype 3 HEV strains prevalent in humans and swine in this area might come from a common infection source.
In the 1980s and early 1990s, genotype 1 was considered the predominant genotype in China; since 2000, genotype 4 HEV has become the dominant cause of hepatitis E disease in China (15). Our current study showed that genotype 3 HEV could be associated with human HEV infection in China. These results may provide a hint that China is transitioning from experiencing HEV infection that was primarily associated with HEV strains transmitted along the human-to-human route to strains transmitted zoonotically.
Dr Zhang is a researcher in School of Medical Science and Laboratory Medicine, Jiangsu University. His primary research interest is the evolution of RNA viruses.
Acknowledgment
This study was supported by the Professional Research Foundation for Advanced Talents of Jiangsu University under grant no.10JDG059.
References
- Reyes GR, Purdy MA, Kim JP, Luk KC, Young LM, Fry KE, Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247:1335–9. DOIPubMedGoogle Scholar
- Meng XJ, Purcell RH, Halbur PG, Lehman JR, Webb DM, Tsareva TS, A novel virus in swine is closely related to the human hepatitis E virus. Proc Natl Acad Sci U S A. 1997;94:9860–5. DOIPubMedGoogle Scholar
- Zheng Y, Ge S, Zhang J, Guo Q, Ng MH, Wang F, Swine as a principal reservoir of hepatitis E virus that infects humans in eastern China. J Infect Dis. 2006;193:1643–9. DOIPubMedGoogle Scholar
- Lu L, Li C, Hagedorn CH. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Rev Med Virol. 2006;16:5–36. DOIPubMedGoogle Scholar
- Ning H, Niu Z, Yu R, Zhang P, Dong S, Li Z. Identification of genotype 3 hepatitis E virus in fecal samples from a pig farm located in a Shanghai suburb. Vet Microbiol. 2007;121:125–30. DOIPubMedGoogle Scholar
- Cooper K, Huang FF, Batista L, Rayo CD, Bezanilla JC, Toth TE, Identification of genotype 3 hepatitis E virus (HEV) in serum and faecal samples from pigs in Thailand and Mexico, where genotype 1 and 2 HEV strains are prevalent in the respective human populations. J Clin Microbiol. 2005;43:1684–8. DOIPubMedGoogle Scholar
- Si FS, Zhu YM, Dong SJ, Yu SS, Yu RS, Shen SY, Full genomic sequence analysis of swine genotype 3 hepatitis E virus isolated from Shanghai. Virus Res. 2009;144:290–3. DOIPubMedGoogle Scholar
- Li RC, Ge SX, Li YP, Zheng YJ, Nong Y, Guo QS, Seroprevalence of hepatitis E virus infection, rural southern People's Republic of China. Emerg Infect Dis. 2006;12:1682–8.PubMedGoogle Scholar
- Yan Y, Zhang W, Shen Q, Cui L, Hua X. Prevalence of four different subgenotypes of genotype 4 hepatitis E virus among swine in the Shanghai area of China. Acta Vet Scand. 2008;50:12. DOIPubMedGoogle Scholar
- Shen Q, Zhang W, Cao X, Mou J, Cui L, Hua X. Cloning of full genome sequence of hepatitis E virus of Shanghai swine isolate using RACE method. Virol J. 2007;4:98. DOIPubMedGoogle Scholar
- Takahashi K, Okamoto H, Abe N, Kawakami M, Matsuda H, Mochida S, Virulent strain of hepatitis E virus genotype 3, Japan. Emerg Infect Dis. 2009;15:704–9. DOIPubMedGoogle Scholar
- Rutjes SA, Lodder WJ, Lodder-Verschoor F, van den Berg HH, Vennema H, Duizer E, Sources of hepatitis E virus genotype 3 in the Netherlands. Emerg Infect Dis. 2009;15:381–7. DOIPubMedGoogle Scholar
- Legrand-Abravanel F, Mansuy JM, Dubois M, Kamar N, Peron JM, Rostaing L, Hepatitis E virus genotype 3 diversity, France. Emerg Infect Dis. 2009;15:110–4. DOIPubMedGoogle Scholar
- Zhang W, Yang S, Shen Q, Huang F, Shan T, Yang Z, Genotype 3 hepatitis E virus existed among swine groups in 4 geographically far regions in China. Vet Microbiol. 2010;140:193–5. DOIPubMedGoogle Scholar
- Wang Y. Epidemiology, molecular biology and zoonosis of genotype IV hepatitis E in China. Chin J Epidemiol. 2003;24:618–22.
Figures
Cite This ArticleTable of Contents – Volume 16, Number 10—October 2010
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Wen Zhang, School of Medical Science and Laboratory Medicine, Jiangsu University, 301 Xuefu Rd, Zhenjiang, Jiangsu 212013, People’s Republic of China
Top