Volume 16, Number 11—November 2010
Research
Measles Virus Strain Diversity, Nigeria and Democratic Republic of the Congo
Figure 1
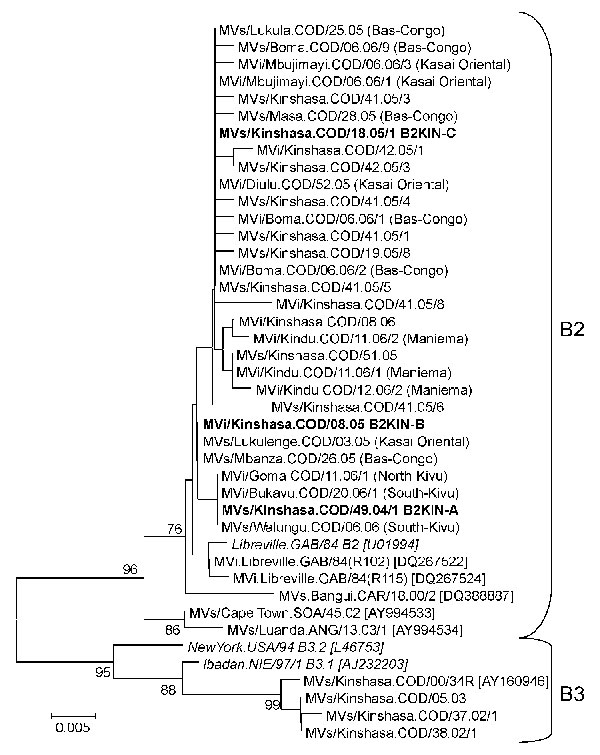
Figure 1. Phylogenetic tree including genotype B2 and genotype B3 of measles virus (MV) strains from the Democratic Republic of the Congo 2000–2006, and World Health Organization (WHO) reference strains (italics) of the corresponding genotypes and some other genotype B2 strains available in GenBank (accession numbers in brackets). MV strains were named according to WHO nomenclature: MVi/City of isolation.Country/epidemiologic week.year of isolation/isolate number. Sequences obtained from RNA extracted from isolates (MVi) or clinical material (MVs) were distinguished. The main genotype B2 variants from Kinshasa (B2KIN-A, B2KIN-B, and B2KIN-C) are indicated in boldface. Except for B2 strains from Kinshasa, the provinces of Democratic Republic of the Congo where strains were collected are indicated in brackets. The phylogenetic tree was calculated on the basis of the 450-nt region that codes for the C-terminus of the MV N protein by using MEGA4 software (24) and the neighbor-joining method (Kimura 2-parameter, 1,000 bootstraps). Scale bar indicates nucleotide substitutions per site.
References
- Otten M, Kezaala R, Fall A, Masresha B, Martin R, Cairns L, Public-health impact of accelerated measles control in the WHO African Region 2000–03. Lancet. 2005;366:832–9. DOIPubMedGoogle Scholar
- Grais RF, Dubray C, Gerstl S, Guthmann JP, Djibo A, Nargaye KD, Unacceptably high mortality related to measles epidemics in Niger, Nigeria, and Chad. PLoS Med. 2007;4:e16. DOIPubMedGoogle Scholar
- Schimmer B, Ihekweazu C. Polio eradication and measles immunisation in Nigeria. Lancet Infect Dis. 2006;6:63–5. DOIPubMedGoogle Scholar
- World Health Organization. Progress in global measles control and mortality reduction, 2000–2006. Wkly Epidemiol Rec. 2007;82:418–24.PubMedGoogle Scholar
- World Health Organization. WHO vaccine-preventable diseases: monitoring system 2006 global summary [cited 2010 Aug 15]. http://www.who.int/vaccines-documents/GlobalSummary/GlobalSummary.pdf
- World Health Organization. Impact of measles control activities in the WHO African Region, 1999–2005. Wkly Epidemiol Rec. 2006;81:365–71.PubMedGoogle Scholar
- Centers for Disease Control and Prevention. Progress toward measles control—African region, 2001–2008. MMWR Morb Mortal Wkly Rep. 2009;58:1036–41.PubMedGoogle Scholar
- Muller CP, Mulders MN. Molecular epidemiology of measles virus. In: Leitner T, editor. The molecular epidemiology of human viruses. Boston: Kluwer Academic Publishers; 2002. p. 237–72.
- Riddell MA, Rota JS, Rota PA. Review of the temporal and geographical distribution of measles virus genotypes in the prevaccine and postvaccine eras. Virol J. 2005;2:87. DOIPubMedGoogle Scholar
- Hanses F, Truong AT, Ammerlaan W, Ikusika O, Adu F, Oyefolu AO, Molecular epidemiology of Nigerian and Ghanaian measles virus isolates reveals a genotype circulating widely in western and central Africa. J Gen Virol. 1999;80:871–7.PubMedGoogle Scholar
- Lemma E, Smit SB, Beyene B, Nigatu W, Babaniyi OA. Genetic characterization and progression of B3 measles genotype in Ethiopia: a study of five measles outbreak cases. Ethiop Med J. 2008;46:79–85.PubMedGoogle Scholar
- Rota J, Lowe L, Rota P, Bellini W, Redd S, Dayan G, Identical genotype B3 sequences from measles patients in 4 countries, 2005. Emerg Infect Dis. 2006;12:1779–81.PubMedGoogle Scholar
- Gouandjika-Vasilache I, Waku-Kouomou D, Menard D, Beyrand C, Guye F, Ngoay-Kossy JC, Cocirculation of measles virus genotype B2 and B3.1 in Central African Republic during the 2000 measles epidemic. J Med Virol. 2006;78:964–70. DOIPubMedGoogle Scholar
- Mbugua FM, Okoth FA, Gray M, Kamau T, Kalu A, Eggers R, Molecular epidemiology of measles virus in Kenya. J Med Virol. 2003;71:599–604. DOIPubMedGoogle Scholar
- Mulders MN, Nebie YK, Fack F, Kapitanyuk T, Sanou O, Valea DC, Limited diversity of measles field isolates after a national immunization day in Burkina Faso: progress from endemic to epidemic transmission? J Infect Dis. 2003;187(Suppl 1):S277–82. DOIPubMedGoogle Scholar
- Kouomou DW, Nerrienet E, Mfoupouendoun J, Tene G, Whittle H, Wild TF. Measles virus strains circulating in Central and West Africa: geographical distribution of two B3 genotypes. J Med Virol. 2002;68:433–40. DOIPubMedGoogle Scholar
- El Mubarak HS, van de Bildt MW, Mustafa OA, Vos HW, Mukhtar MM, Ibrahim SA, Genetic characterization of wild-type measles viruses circulating in suburban Khartoum, 1997–2000. J Gen Virol. 2002;83:1437–43.PubMedGoogle Scholar
- Muwonge A, Nanyunja M, Rota PA, Bwogi J, Lowe L, Liffick SL, New measles genotype, Uganda. Emerg Infect Dis. 2005;11:1522–6.PubMedGoogle Scholar
- Rota PA, Bloom AE, Vanchiere JA, Bellini WJ. Evolution of the nucleoprotein and matrix genes of wild-type strains of measles virus isolated from recent epidemics. Virology. 1994;198:724–30. DOIPubMedGoogle Scholar
- Rota PA, Featherstone DA, Bellini WJ. Molecular epidemiology of measles virus. Curr Top Microbiol Immunol. 2009;330:129–50. DOIPubMedGoogle Scholar
- World Health Organization. Manual for the laboratory diagnosis of measles and rubella infection. Geneva: The Organization; 2006.
- Santibanez S, Tischer A, Heider A, Siedler A, Hengel H. Rapid replacement of endemic measles virus genotypes. J Gen Virol. 2002;83:2699–708.PubMedGoogle Scholar
- Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002;Chapter 2:Unit 2.3. Medline
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
- Grais RF, Ferrari MJ, Dubray C, Fermon F, Guerin PJ. Exploring the time to intervene with a reactive mass vaccination campaign in measles epidemics. Epidemiol Infect. 2006;134:845–9. DOIPubMedGoogle Scholar
- Ferrari MJ, Grais RF, Bharti N, Conlan AJ, Bjornstad ON, Wolfson LJ, The dynamics of measles in sub-Saharan Africa. Nature. 2008;451:679–84. DOIPubMedGoogle Scholar
- Gay NJ. The theory of measles elimination: implications for the design of elimination strategies. J Infect Dis. 2004;189(Suppl 1):S27–35. DOIPubMedGoogle Scholar
- Nojiri S, Vynnycky E, Gay N. Interpreting changes in measles genotype: the contribution of chance, migration and vaccine coverage. BMC Infect Dis. 2008;8:44. DOIPubMedGoogle Scholar
- Zhang Y, Zhen Z, Rota PA, Jiang X, Hu J, Wang J, Molecular epidemiology of measles viruses in China 1995–2003. Virol J. 2007;4:14. DOIPubMedGoogle Scholar
- Anderson RM, May RM. Directly transmitted infections diseases: control by vaccination. Science. 1982;215:1053–60. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Update: global measles control and mortality reduction–worldwide, 1991–2001. MMWR Morb Mortal Wkly Rep. 2003;52:471–5.PubMedGoogle Scholar
- Dubray C, Gervelmeyer A, Djibo A, Jeanne I, Fermon F, Soulier MH, Late vaccination reinforcement during a measles epidemic in Niamey, Niger (2003–2004). Vaccine. 2006;24:3984–9. DOIPubMedGoogle Scholar
- Grais RF, Conlan AJ, Ferrari MJ, Djibo A, Le Menach A, Bjornstad ON, Time is of the essence: exploring a measles outbreak response vaccination in Niamey, Niger. J R Soc Interface. 2008;5:67–74. DOIPubMedGoogle Scholar
- Grais RF, Ferrari MJ, Dubray C, Bjornstad ON, Grenfell BT, Djibo A, Estimating transmission intensity for a measles epidemic in Niamey, Niger: lessons for intervention. Trans R Soc Trop Med Hyg. 2006;100:867–73. DOIPubMedGoogle Scholar
- Smit SB, Hardie D, Tiemessen CT. Measles virus genotype B2 is not inactive: evidence of continued circulation in Africa. J Med Virol. 2005;77:550–7. DOIPubMedGoogle Scholar