Volume 16, Number 9—September 2010
Dispatch
Novel Hepatitis E Virus Genotype in Norway Rats, Germany
Figure A1
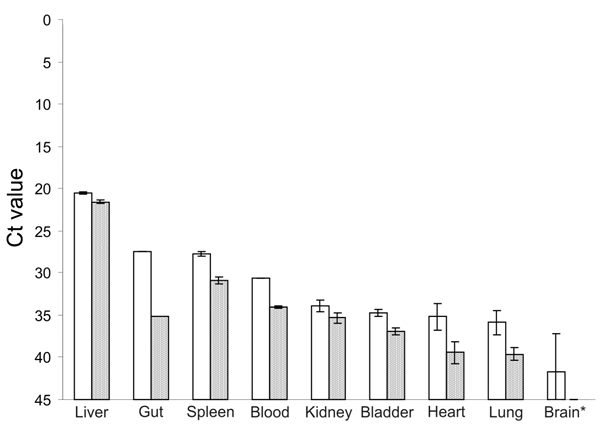
Figure A1. Results of real-time reverse transcription–PCR showing cycle threshold (Ct) values of viral load in different tissues of Norway rats nos. 63 (white bars) and 68 (gray bars). For RNA isolation, 10 mg of tissue or 10 µL of blood were homogenized and used. The PCR was selective for a region in the open reading frame 2 of rat hepatitis E virus (rHEV) and was based on primers rHEV-forward (5′-TACCCGATGCCGGGCAGT-3′) and rHEV-reverse (5′-ATCCACATCTGGGACAGG-3′) and probe (5′-6FAM-AATGACAGCACAGGCACC-BBQ-3′). Error bars indicate SD. *Brain sample from rat no. 63 contained blood.
Page created: August 28, 2011
Page updated: August 28, 2011
Page reviewed: August 28, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.