Volume 17, Number 10—October 2011
Dispatch
Novel Amdovirus in Gray Foxes
Figure 1
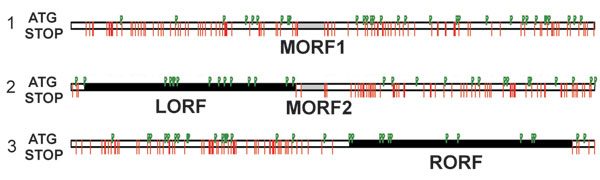
Figure 1. Open reading frames (ORFs) in gray fox amdovirus genome. Three possible reading frames of the plus-strand sequence with the stop codons indicated by red lines and ATG codons by green flags. Two major ORFs, left (LORF) and right (RORF), are indicated by black bars; 2 small middle ORFs (MORF1 and MORF2) are indicated by gray bars.
Page created: September 26, 2011
Page updated: September 26, 2011
Page reviewed: September 26, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.