Volume 17, Number 4—April 2011
Research
Genomic Analysis of Highly Virulent Georgia 2007/1 Isolate of African Swine Fever Virus
Figure 1
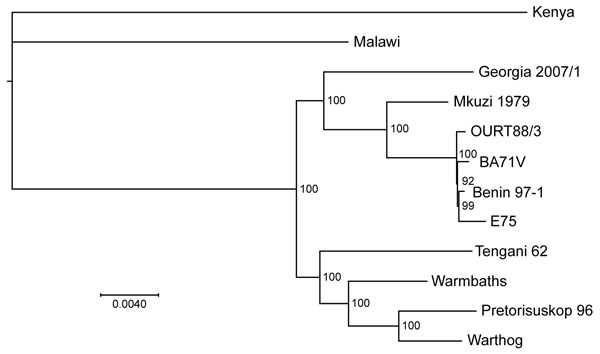
Figure 1. Comparison of the Georgia 2007/1 African swine fever virus (ASFV) isolate genome with those of other ASFV isolates. ASFV phylogeny midpoint was rooted in a neighbor-joining tree on the basis of 125 conserved open reading frame regions (40,810 aa) from 12 taxa. Node values show percentage bootstrap support (n = 1,000). The isolates shown and accession numbers are Kenya AY261360, Malawi Lil20/1 AY261361, Tengani AY261364, Warmbaths AY261365, Pretorisuskop AY261363, Warthog AY261366, Warmbaths AY261365, Mkuzi AY261362, OurT88/3 a.m.712240, BA71V NC_001659, Benin97/1 a.m.712239, and E75 FN557520. Scale bar indicates nucleotide substitutions per site.
Page created: July 25, 2011
Page updated: July 25, 2011
Page reviewed: July 25, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.