Volume 17, Number 7—July 2011
Letter
Plasmodium knowlesi Reinfection in Human
Figure
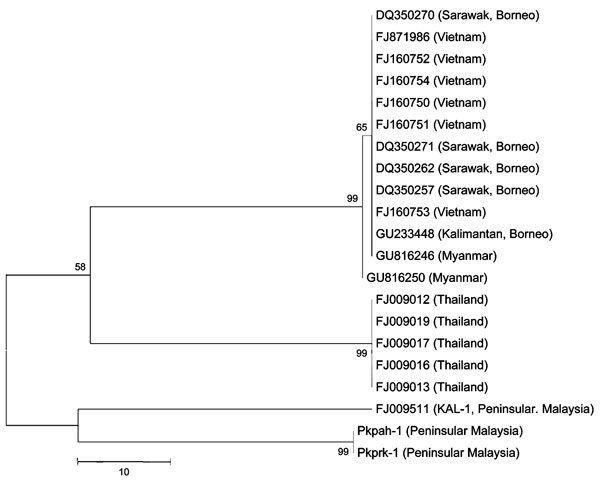
Figure. Phylogenetic trees based on nucleotide sequences of small subunit rRNA of Plasmodium knowlesi isolates from Peninsular Malaysia (Pkpah-1, Pkprk-1, KAL-1) and surrounding regions (denoted by GenBank accession nos.). The tree was constructed by using the maximum-parsimony method. The percentage of replicate trees in which the associated isolates cluster together in the bootstrap test (10,000 and 1,000 replicates, no differences were observed) is shown next to the branches. Phylogenetic analysis was conducted by using MEGA4 (6). Scale bar indicates nucleotide substitutions per site.
References
- Singh B, Lee KS, Matusop A, Radhakrishnan A, Shamsul SSG, Cox-Singh J, A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–24. DOIPubMedGoogle Scholar
- Vythilingam I, Noorazian YM, Huat TC, Jiram AI, Yusri YM, Azahari AH, Plasmodium knowlesi in humans, macaques and mosquitoes in Peninsular Malaysia. Parasit Vectors. 2008;1:26. DOIPubMedGoogle Scholar
- Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–92.PubMedGoogle Scholar
- Daneshvar C, Davis C, Cox-Singh J, Rafa'ee M, Zakaria S, Divis P, Clinical and laboratory features of human Plasmodium falciparum infections. Clin Infect Dis. 2009;49:852–60. DOIPubMedGoogle Scholar
- McCutchan TF, Kissinger JC, Touray MG, Rogers MJ, Li J, Sullivan M, Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications. Proc Natl Acad Sci U S A. 1996;93:11889–94. DOIPubMedGoogle Scholar
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
- Kantele A, Marti H, Felger I, Müller D, Jokiranta TS. Monkey malaria in a European traveler returning from Malaysia. Emerg Infect Dis. 2008;14:1434–6. DOIPubMedGoogle Scholar
- Krotoski WA, Collins WE. Failure to detect hypnozoites in hepatic tissue containing exoerythrocytic schizonts of Plasmodium knowlesi. Am J Trop Med Hyg. 1982;31:854–6.PubMedGoogle Scholar
- A single parasite gene determines strain-specific protective immunity against malaria: the role of the merozoite surface protein I. Int J Parasitol. 2010;40:951–61. DOIPubMedGoogle Scholar
Page created: August 15, 2011
Page updated: August 15, 2011
Page reviewed: August 15, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.