Volume 17, Number 8—August 2011
Dispatch
Aichi Virus Shedding in High Concentrations in Patients with Acute Diarrhea
Figure 2
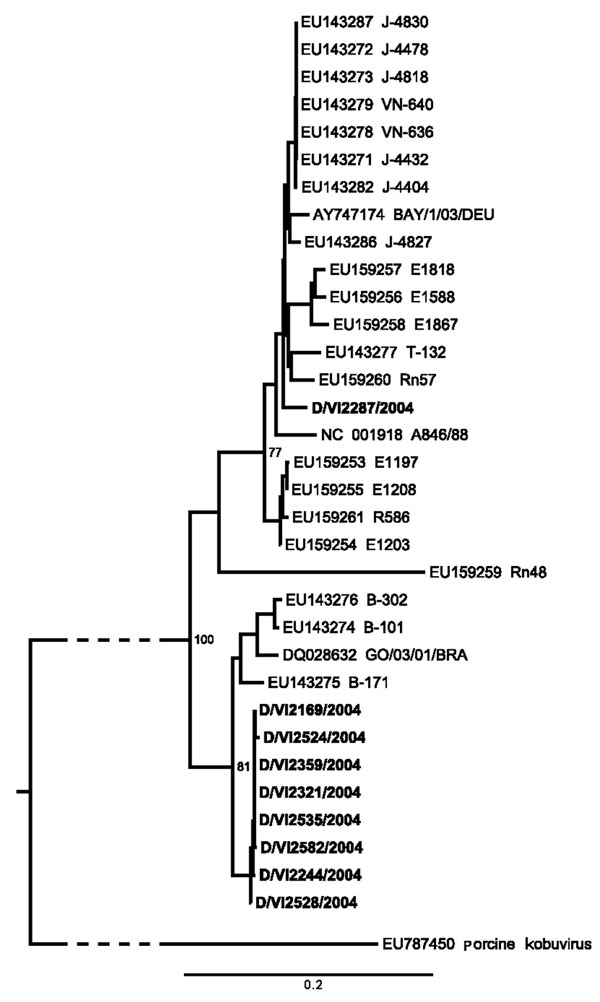
Figure 2. Neighbor-joining phylogeny of Aichi virus (AiV) viral protein 1 gene of strains from study of AiV in patients with acute diarrhea (boldface), Germany, compared with strains from GenBank. The tree was generated by using MEGA4 (www.megasoftware.net) using the maximum-composite likelihood nucleotide substitution model and complete deletion option. Porcine kobuvirus was used as an outgroup (branch truncated as indicated by slashed lines). Bootstrap values from 1,000 reiterations are depicted next to root points. The final dataset corresponded to nucleotide positions 3,034–3,663 in AiV GenBank accession no. AB040749. Scale bar indicates number of base substitutions per site.
Page created: August 15, 2011
Page updated: August 15, 2011
Page reviewed: August 15, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.