Volume 17, Number 8—August 2011
Research
Human Polyomavirus Related to African Green Monkey Lymphotropic Polyomavirus
Figure 3
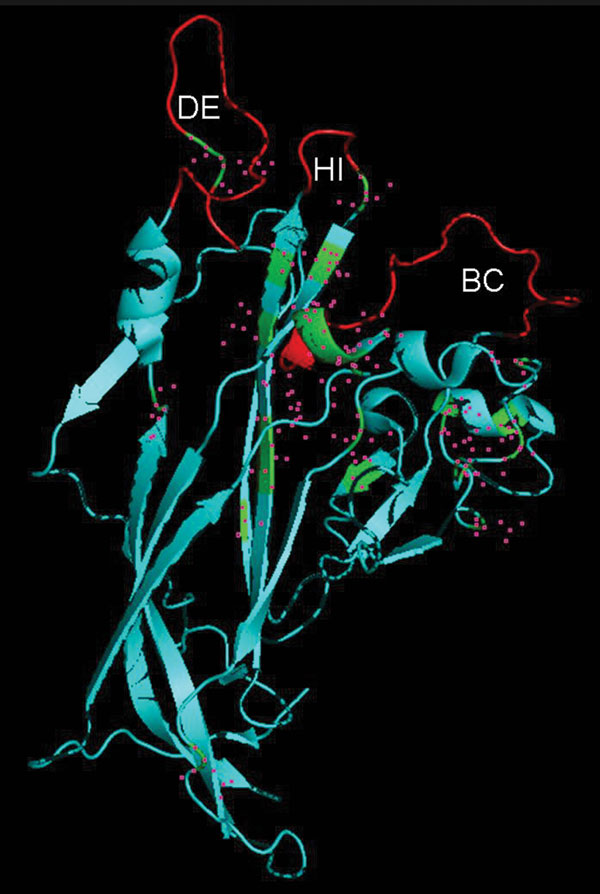
Figure 3. Identification of viral protein 1 (VP1) residues differing between human polyomavirus 9 (HPyV9) and lymphotropic polyomavirus (LPV). The DE, HI, and BC loops that extend outward from VP1 are indicated. The crystal structure of simian virus VP1, derived from strain 3BWQ, was used as a template. The red region in the center indicates part of a β strand, which is mostly hidden. Residues differing between HPyV9 and LPV are indicated by pink squares.
Page created: August 16, 2011
Page updated: August 16, 2011
Page reviewed: August 16, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.