Volume 18, Number 3—March 2012
Research
Lineage-specific Virulence Determinants of Haemophilus influenzae Biogroup aegyptius
Figure 2
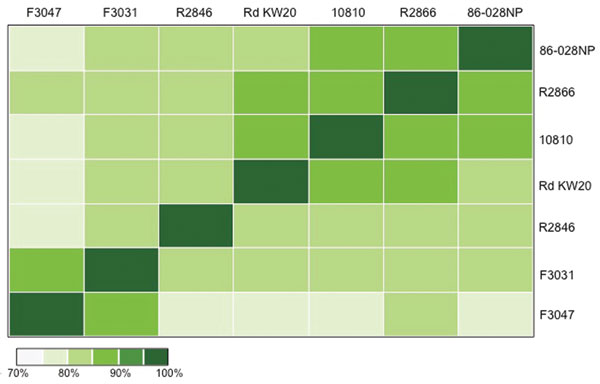
Figure 2. Pair-wise comparisons of genome alignments between 7 Haemophilus influenzae strains. Each colored block represents the total number of bases shared between 2 H. influenzae genomes. Scale bar indicates percent relatedness.
1Current affiliation: Stanford University, Palo Alto, California, USA.
Page created: February 16, 2012
Page updated: February 16, 2012
Page reviewed: February 16, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.