Volume 18, Number 6—June 2012
Letter
Novel Chlamydiaceae Disease in Captive Salamanders
Figure
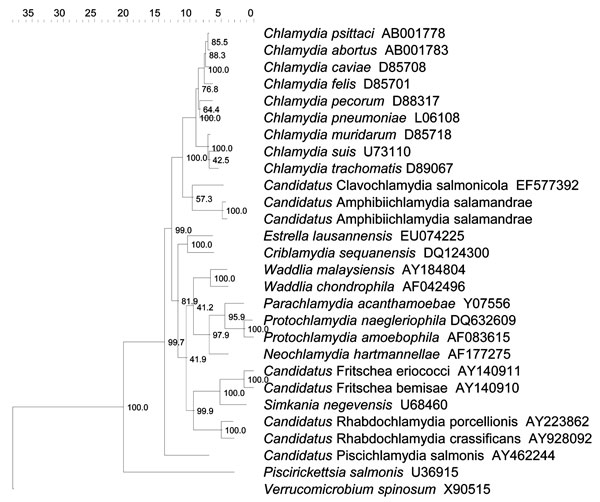
Figure. . . Topology of the novel amphibian Chlamydiaceae (Candidatus Amphibiichlamydia salamandrae) within the phylogenetic tree obtained by neighbor-joining and based on 16S rRNA gene data from representative species. Numbers show the percentage of times each branch was found in 1,000 bootstrap replicates. The tree has been rooted with Verrucomicrobium spinosum as outgroup. Scale bar indicates nucleotide substitutions per site.
Page created: May 16, 2012
Page updated: May 16, 2012
Page reviewed: May 16, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.