Volume 19, Number 1—January 2013
Dispatch
Novel Epidemic Clones of Listeria monocytogenes, United States, 2011
Figure 1
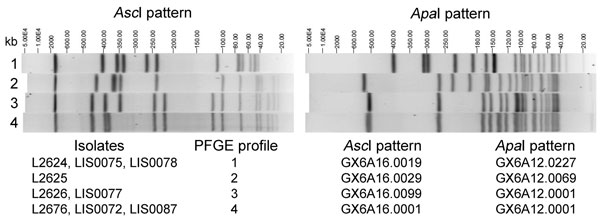
Figure 1. . . Four AscI/ApaI pulsed-field gel electrophoresis (PFGE) profiles (identified at the time the research was performed) displayed by Listeria monocytogenes clinical isolates (L2624, L2625, L2626, and L2676) and isolates from food or environmental samples (LIS0072, LIS0075, LIS0077, LIS0078, and LIS0087) associated with the 2011 listeriosis outbreak traced to cantaloupe. PFGE profiles 3 and 4 differ by ≈40-kb shift in 1 band in the AscI pattern, likely related to the loss or acquisition of the comK prophage, because the size of this prophage was ≈40 kb as calculated by using the whole genome sequencing data (not shown).
Page created: December 20, 2012
Page updated: December 20, 2012
Page reviewed: December 20, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.