Volume 20, Number 5—May 2014
Research
Molecular Characterization of Cryptically Circulating Rabies Virus from Ferret Badgers, Taiwan
Figure 5
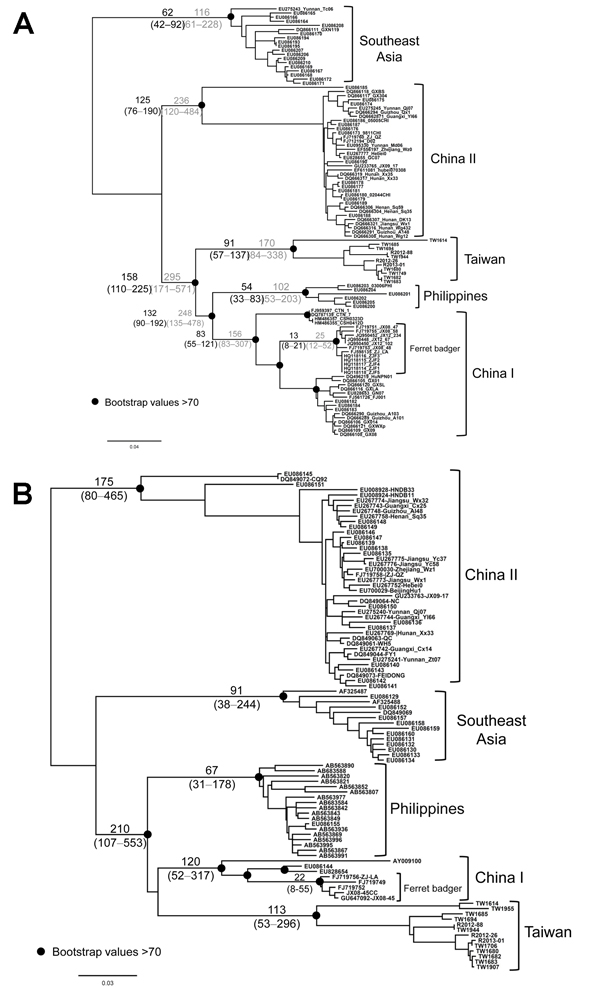
Figure 5. Maximum-likelihood trees of rabies virus based on (A) nucleoprotein and (B) glycoprotein gene sequencesNumbers on the branches are estimated divergences (above) and their 95% highest posterior density (below)The divergence time between different viral lineages and time to the most recent common ancestor of virus isolates were estimated by using an established Bayesian Markov chain Monte Carlo approach implemented in BEAST version 1.7 (21)The substitution rate was assumed to be 4.3 × 10–4 (range 3.1–6.6 × 10–4) or 2.3 × 10–4 (range 1.1–3.6 × 10–4)/sites/year (hatched numbers) for nucleoprotein (N) and 3.9 × 10–4 (1.2–6.5 × 10–4)/sites/year for glycoprotein (G) genes (17,23)The analysis was performed by using the general time-reversible model of nucleotide substitution, assuming an uncorrelated lognormal molecular clock (22)TW1955 is the only isolate from a shrew that is very close to TW1614 collected from the same area, suggesting that TW1955 was a spillover from a ferret badger (see also Figure 1)Scale bar indicates nucleotide substitutions per site.
References
- Jackson AC. Pathogenesis. In: Jackson AC, Wunner WH, editors. Rabies. London: Elsevier Academic Press; 2007.
- World Health Organization. WHO Expert Consultation on Rabies: second report. Geneva. Organization. 2013;•••:1–139.
- Wandeler A. Virus infections of non-domestic carnivores: rabies virus. In: Appel MJ, editor. Virus infections of carnivores. Amsterdam: Elsevier Science Publishers; 1987. p. 449–61.
- Barnard BJH. The role played by wildlife in the epizootiology of rabies in South Africa and South-West Africa. Onderstepoort J Vet Res. 1979;46:155–63 .PubMedGoogle Scholar
- Smith GC, Wilkinson D. Modelling disease spread in a novel host: rabies in the European badger Meles meles. J Appl Ecol. 2002;39:865–74. DOIGoogle Scholar
- Wandeler A, Wachendorfer G, Forster U, Krekel H, Muller J, Steck F. Rabies in wild carnivores in central Europe. II. Virological and serological examinations. Zentralbl Veterinarmed B. 1974;21:757–64 . DOIPubMedGoogle Scholar
- Liu Y, Zhang SF, Wu XF, Zhao JH, Hou YL, Zhang F, Ferret badger rabies origin and its revisited importance as potential source of rabies transmission in southeast China. BMC Infect Dis. 2010;10:234. DOIPubMedGoogle Scholar
- Zhenyu G, Zhen W, Enfu C, Fan H, Junfen L, Yixin L, Human rabies cluster following badger bites, People’s Republic of China. Emerg Infect Dis. 2007;13:1956–7. DOIPubMedGoogle Scholar
- Zhang S, Tang Q, Wu XF, Liu Y, Zhang F, Rupprecht CE, Rabies in ferret badgers, southeastern China. Emerg Infect Dis. 2009;15:946–9 . DOIPubMedGoogle Scholar
- Lei YL, Wang XG, Liu FM, Chen XY, Ye BF, Mei JH, Complete genome sequencing and analyses of rabies viruses isolated from wild animals (Chinese ferret-badger) in Zhejiang Province [in Chinese]. Zhonghua Liu Xing Bing Xue Za Zhi. 2009;30:824–8 .PubMedGoogle Scholar
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. DOIPubMedGoogle Scholar
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. DOIPubMedGoogle Scholar
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95 .PubMedGoogle Scholar
- Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709 .PubMedGoogle Scholar
- He CQ, Meng SL, Yan HY, Ding NZ, He HB, Yan JX, Isolation and identification of a novel rabies virus lineage in China with natural recombinant nucleoprotein gene. PLoS ONE. 2012;7:e49992. DOIPubMedGoogle Scholar
- Bourhy H, Reynes JM, Dunham EJ, Dacheux L, Larrous F, Huong VT, The origin and phylogeography of dog rabies virus. J Gen Virol. 2008;89:2673–81. DOIPubMedGoogle Scholar
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. DOIPubMedGoogle Scholar
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–6. DOIPubMedGoogle Scholar
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–4. DOIPubMedGoogle Scholar
- Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29:1969–73. DOIPubMedGoogle Scholar
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. DOIPubMedGoogle Scholar
- Meng S, Sun Y, Wu X, Tang J, Xu G, Lei Y, Evolutionary dynamics of rabies viruses highlights the importance of China rabies transmission in Asia. Virology. 2011;410:403–9. DOIPubMedGoogle Scholar
- Kissi B, Tordo N, Bourhy H. Genetic polymorphism in the rabies virus nucleoprotein gene. Virology. 1995;209:526–37. DOIPubMedGoogle Scholar
- Mebatsion T, Weiland F, Conzelmann KK. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J Virol. 1999;73:242–50 .PubMedGoogle Scholar
- Finke S, Mueller-Waldeck R, Conzelmann KK. Rabies virus matrix protein regulates the balance of virus transcription and replication. J Gen Virol. 2003;84:1613–21. DOIPubMedGoogle Scholar
- Kassis R, Larrous F, Estaquier J, Bourhy H. Lyssavirus matrix protein induces apoptosis by a TRAIL-dependent mechanism involving caspase-8 activation. J Virol. 2004;78:6543–55. DOIPubMedGoogle Scholar
- Lentz TL, Wilson PT, Hawrot E, Speicher DW. Amino acid sequence similarity between rabies virus glycoprotein and snake venom curaremimetic neurotoxins. Science. 1984;226:847–8. DOIPubMedGoogle Scholar
- Poch O, Blumberg BM, Bougueleret L, Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol. 1990;71:1153–62. DOIPubMedGoogle Scholar
- Kuzmin IV, Wu X, Tordo N, Rupprecht CE. Complete genomes of Aravan, Khujand, Irkut and West Caucasian bat viruses, with special attention to the polymerase gene and non-coding regions. Virus Res. 2008;136:81–90. DOIPubMedGoogle Scholar
- Zhang S, Zhao J, Liu Y, Fooks AR, Zhang F, Hu R. Characterization of a rabies virus isolate from a ferret badger (Melogale moschata) with unique molecular differences in glycoprotein antigenic site III. Virus Res. 2010;149:143–51. DOIPubMedGoogle Scholar
- Gong W, Jiang Y, Za Y, Zeng Z, Shao M, Fan J, Temporal and spatial dynamics of rabies viruses in China and Southeast Asia. Virus Res. 2010;150:111–8. DOIPubMedGoogle Scholar
- Saito M, Oshitani H, Orbina JR, Tohma K, de Guzman AS, Kamigaki T, Genetic diversity and geographic distribution of genetically distinct rabies viruses in the Philippines. PLoS Negl Trop Dis. 2013;7:e2144. DOIPubMedGoogle Scholar
- Streicker DG, Lemey P, Velasco-Villa A, Rupprecht CE. Rates of viral evolution are linked to host geography in bat rabies. PLoS Pathog. 2012;8:e1002720. DOIPubMedGoogle Scholar
- Biek R, Henderson JC, Waller LA, Rupprecht CE, Real LA. A high-resolution genetic signature of demographic and spatial expansion in epizootic rabies virus. Proc Natl Acad Sci U S A. 2007;104:7993–8. DOIPubMedGoogle Scholar
- Faber M, Faber ML, Papaneri A, Bette M, Weihe E, Dietzschold B, A single amino acid change in rabies virus glycoprotein increases virus spread and enhances virus pathogenicity. J Virol. 2005;79:14141–8. DOIPubMedGoogle Scholar
- Faber M, Li J, Kean RB, Hooper DC, Alugupalli KR, Dietzschold B. Effective preexposure and postexposure prophylaxis of rabies with a highly attenuated recombinant rabies virus. Proc Natl Acad Sci U S A. 2009;106:11300–5. DOIPubMedGoogle Scholar
- Tuffereau C, Schmidt K, Langevin C, Lafay F, Dechant G, Koltzenburg M. The rabies virus glycoprotein receptor p75NTR is not essential for rabies virus infection. J Virol. 2007;81:13622–30. DOIPubMedGoogle Scholar
- Kuzmin IV, Shi M, Orciari LA, Yager PA, Velasco-Villa A, Kuzmina NA, Molecular inferences suggest multiple host shifts of rabies viruses from bats to mesocarnivores in Arizona during 2001–2009. PLoS Pathog. 2012;8:e1002786 . DOIPubMedGoogle Scholar
1Joint senior authors who contributed equally to this article.