Volume 25, Number 1—January 2019
Research
Association of Increased Receptor-Binding Avidity of Influenza A(H9N2) Viruses with Escape from Antibody-Based Immunity and Enhanced Zoonotic Potential
Abstract
We characterized 55 influenza A(H9N2) viruses isolated in Pakistan during 2014–2016 and found that the hemagglutinin gene is of the G1 lineage and that internal genes have differentiated into a variety of novel genotypes. Some isolates had up to 4-fold reduction in hemagglutination inhibition titers compared with older viruses. Viruses with hemagglutinin A180T/V substitutions conveyed this antigenic diversity and also caused up to 3,500-fold greater binding to avian-like and >20-fold greater binding to human-like sialic acid receptor analogs. This enhanced binding avidity led to reduced virus replication in primary and continuous cell culture. We confirmed that altered receptor-binding avidity of H9N2 viruses, including enhanced binding to human-like receptors, results in antigenic variation in avian influenza viruses. Consequently, current vaccine formulations might not induce adequate protective immunity in poultry, and emergence of isolates with marked avidity for human-like receptors increases the zoonotic risk.
Since their first detection in China in 1992, avian influenza A(H9N2) viruses of the G1 and BJ94 lineages have become enzootic to poultry in Asia and parts of Africa (1–3). These viruses frequently cause outbreaks in these regions and sporadic outbreaks in Europe and North America (4–9). H9N2 viruses cause moderate illness and death rates in domestic poultry, leading to major economic burden to small-scale and large-scale poultry industries, and increased risk for zoonotic infection (6,10,11).
Severe illness in humans infected with this virus is rare, but seroepidemiologic data suggest that infection might be most common in those working at the human–animal interface (12–14). It is evident that there is a major genetic host barrier between currently circulating H9N2 viruses and humans, despite detection of molecular markers of mammalian tropism in avian isolates (15,16). Adaptation to humans requires permissive mutations throughout the genome of avian influenza viruses that affect multiple factors, such as receptor binding, pH stability, virus polymerase activity, innate immune responses, and viral egress (17–21).
Hemagglutinin (HA) and neuraminidase (NA) play critical roles in overcoming this genetic host barrier, as shown by recent zoonotic infections with H7N9, H10N8, and H5Nx viruses, all containing H9N2 internal genes, compared with the remarkable dearth of reported human infections with H9N2 viruses, despite their higher incidence in poultry (15,22,23). Efficient virus replication is dependent, in part, on the concerted activities of HA and NA binding and eluting cells and is a balance maintained by matching HA substitutions that alter receptor-binding avidity with changes to NA stalk length and sialidase activity (24–26).
HA is a membrane-bound homotrimer that becomes functionally active through proteolytic cleavage of precursor HA0 into HA1 and HA2 polypeptides. The HA1 globular head domain facilitates host-cell binding, which initiates infection, and the HA2 fusion peptide, primed during cleavage activation, facilitates pH-dependent fusion between virus and host-cell membranes (27). Human-to-human transmissible influenza viruses typically display preferential binding to glycans containing terminal α2,6-linked sialic acids (SAs), which are prevalent in the upper respiratory tract of humans (19,28,29); avian-origin viruses typically bind preferentially to glycans having terminal α2,3-linked SAs, which are found throughout the avian gastrointestinal and respiratory tracts (30). The 3′-sialyllactosamine and 6′-sialyllactosamine receptor analogs are commonly used in influenza receptor-binding assays representing α2,3-linked (3SLN) and α2,6-linked (6SLN) SAs (31). The switch in binding preference of H9N2 viruses from avian-like α2,3-linked to human-like α2,6-linked SAs has been attributed to amino acid substitutions in the receptor-binding site (RBS) of HA1, with residue 216 [226] (mature H9 numbering used throughout; H3 numbering within brackets) frequently shown to play a critical role (32–34).
Increasing evidence suggests that the combination and nature of amino acids at additional positions (e.g., 180 [190] and 217 [227]) needs to be determined for assessing receptor binding and zoonotic potential of H9 subtype viruses (17). H9N2 viruses commonly have alanine at HA1 position 180 (A180), which correlates with a preference for binding of 3SLN, viruses with glutamic acid (E180) generally show greater binding avidity for 6SLN; and viruses with valine (V180) have been shown to show appreciable binding to 6SLN, together with greater replicative fitness in mammals than viruses with A180 (17,35,36).
The HAs of H9N2 viruses in Pakistan typically contain the RBS residues A180, L216, and I217. A/chicken/Pakistan/UDL-01/2008 (UDL-01/08) and A/chicken/Pakistan/UDL-02/2008 were shown to preferentially bind a variant of the 3SLN receptor analog, which is sulfated at the 6′ position of the penultimate sugar Neu5Ac α2,3 β1–4(6-HSO3)GlcNAc (3SLN(6Su)). Although these viruses contain the classical humanizing residue L216, they show negligible binding to 6SLN in quantitative receptor-binding assays (16,17).
For this study, we conducted influenza surveillance for poultry farms in Pakistan during 2014–2016. Since the late 1990s, H9N2 virus has become enzootic in Pakistan (37). In addition, periodic outbreaks of infection with highly pathogenic avian influenza A(H5N1) and A(H7N3) viruses during 1995–2006 have occurred, resulting in intersubtype reassortment events and new viruses, some of which have subsequently spread to become predominant virus strains in the region (9,16,38). We aimed to characterize the genetics and antigenicity of circulating H9N2 viruses in Pakistan and link the molecular basis of differences seen in circulating viruses with zoonotic potential. Specifically, we sought to show that emergence of substitutions at HA1 position 180 are driving increases in virus receptor-binding avidity, including for human-like receptor analogs, and concurrently enabling virus to escape antibody-based immunity (Appendix).
Virus Isolation and Phylogenetic Analysis
We collected 1,374 oropharyngeal and cloacal swab specimens from poultry during surveillance of farms for avian influenza viruses in Pakistan during 2014–2016. Of these specimens, 78 (5.7%) were confirmed by hemagglutinin inhibition (HI) assay to be positive for H9, and sequencing data for 55 virus isolates were generated. Next-generation sequencing of isolates yielded 43 complete genomes and 12 partial genomes. No other avian influenza virus subtypes were detected in this study.
Similar to previous results for H9N2 viruses from Pakistan, phylogenetic analysis showed that all HA and NA genes from specimens collected during 2014–2016 continued to belong to the G1 lineage, and all HA genes clustered within the Middle East B clade. Viruses isolated during this study show continued evolution, as shown by phylogenies that demonstrated drift of subclades away from previous isolates (Appendix Figures 1–3). BLASTn (https://blast.ncbi.nlm.nih.gov/Blast.cgi) comparisons for each gene segment of the 2014–2016 viruses against National Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/) and GISAID (https://www.gisaid.org/) databases showed the closest related viruses to be previous isolates from Pakistan, which had >95% nucleotide homology, suggesting continuous in situ evolution.
Comparison of maximum pairwise nucleotide differences between viruses isolated during 2014–2016 showed variable genetic diversity within each gene segment. Segment 8 (nonstructural) showed the greatest diversity (7% nucleotide difference), and segment 5 (nucleoprotein) showed the least diversity (2% nucleotide difference). In many instances, this diversity could be attributed to a small subset of sequenced viruses: when these segments were removed from the pairwise comparison, virus diversity was reduced. Diversity of gene segments of these outlier viruses that was affected (Appendix Figures 1–3) for gene segments affected from 7% to 2.6% for nonstructural, from 6.1% to 1.1% for polymerase basic 2, and from 6.4% to 2% for HA. Diversity of gene segments remained constant even after removal of those outlier viruses from the pairwise comparison: polymerase basic 1, 4.5%; polymerase acidic, 4.5%; nucleoprotein, 2%; NA, 4.8%; and matrix, 5.5%.
It has previously been shown that H9N2 viruses in Pakistan are reassortants between G1 lineage H9N2 and either H7N3 or clade 2.2 H5N1 subtype viruses (16). All internal genes of viruses isolated in this study clustered with the H7N3 virus isolate A/chicken/Pakistan/NARC-100/2004, indicating that this genotype had become predominant in Pakistan. We found no evidence of further intersubtype reassortment.
Using viruses for which we had full-genome sequences and a >2% nt difference cutoff for each segment, we found that viruses isolated during 2014–2016 could be divided into 7 distinct genotypes (Appendix Figures 1–3). Genotypes PK1 and PK2 showed variable intrasubtype reassortment between H9N2 viruses circulating in Pakistan, as shown by phylogenetic incongruence for some genes. Similar patterns of reassortment have been found in Vietnam and China (15,39). PK3 showed less intrasubtype reassortment than PK1 and PK2.
Molecular Characteristics of H9N2 Viruses Isolated during 2014–2016
Influenza virus HA requires activation by host proteases at a conserved cleavage site between the HA1 and HA2 subunits. This site is a key determinant of pathogenicity in avian species and describes the switch between low pathogenicity and highly pathogenic avian influenza viruses. Low pathogenicity avian influenza viruses contain monobasic cleavage sites, and highly pathogenic avian influenza viruses contain polybasic cleavage sites (40). All H9 HA cleavage sites we sequenced were either KSNR/GLF (10/55) or KSSR/GLF (45/55); viruses containing KSNR/GLF grouped in the PK3 genotype. Previously, HA of H9N2 viruses from Pakistan could be separated into 2 groups containing either KSSR/GLF or RSSR/GLF cleavage sites (16), indicating that the KSSR motif has persisted and undergone some evolution to KSNR. These motifs are dibasic and unlikely to be susceptible to activation by endogenous furin-like proteases, leading to classification of these viruses as low pathogenicity avian influenza viruses. However, such dibasic motifs have been described as being susceptible to an extended group of proteases present in a wider range of tissues, potentially enabling these viruses to show increased tissue tropism and pathogenicity (41).
All viruses we sequenced in this study contained HA L216, which has been associated with mammalian tropism of H9N2 viruses (32). However, recent evidence suggests that L216 alone is a poor marker for human-like receptor-binding preference in H9N2 viruses without considering surrounding amino acids, notably at positions 180 and 217 (17). All viruses sequenced contained I217; 52 viruses contained A180 and 3 contained T180.
Antigenic Characterization of H9N2 Viruses from Pakistan
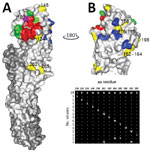
To assess antigenic diversity of the 2014–2016 H9N2 virus subtype population (referred to as contemporary viruses), we compared amino acid sequences of HA to identify substitutions in the surface of HA1 (Figure 1). We cloned HA of a 2016 isolate (SKP-827/16) and used it as a backbone to introduce amino acid substitutions by site-directed mutagenesis, which represented the surface diversity of HA1 of the viruses isolated during 2014–2016. Viruses rescued by reverse genetics (RG) differed by only 1 or 2 aa substitutions that reflected diversity seen between individual sequenced isolates. We then assessed HI titers of viruses isolated during 2014–2016 by using chicken postinfection polyclonal antiserum raised against the closely related Pakistan 2008 isolate UDL-01/08. Most viruses were inhibited at titers equivalent to that with the homologous UDL-01/08 virus, but we observed 4-fold reductions in HI titers for viruses containing the HA A180T substitution (Table 1). We observed the same effect when the HA A180T substitution was introduced into wild-type UDL-01/08. Wild-type SKP-827/16 naturally has HA T180, and a 4-fold increase in microneutralization (MNT) titer with antiserum raised against UDL-01/08 was seen when the T180A substitution was introduced, again indicating that this residue alone was responsible for most antigenic diversity in the sequenced viruses. Conversely, for the UDL-01/08 virus, the A180T substitution caused a 3-fold reduction in MNT titer compared with the titer that had the homologous virus (Table 2).
V180 has also been identified as a potential modulator of receptor binding (35) because the HA A180T substitution caused a major reduction in HI titer with antiserum raised against UDL-01/08. Thus, we assessed whether the A180V substitution had the same effect. Introduction of the HA V180 substitution into UDL-01/08 and SKP-827/16 RG viruses caused 4-fold greater reductions in HI titer by antiserum raised against UDL-01/08 than for parental RG viruses (Table 1). In a similar fashion, we found a 3-fold greater reduction in MNT titer for virus SKP-827/16 containing the T180V substitution and a >6-fold greater reduction in MNT titer for virus UDL-01/08 containing the A180V substitution than for titers with respective parental RG viruses (Table 2). Our results showed that A180V and A180T substitutions were sufficient to produce virus neutralization escape variants, as assessed by HI and MNT assays, when using postinfection polyclonal antiserum raised against UDL-01/08, which contains HA A180 and which recognizes epitopes in 2 discrete antigenic sites (42).
Determination of Receptor-Binding Avidity by Residue 180
The HA RBS is located on the head domain of HA1 and is responsible for recognition of sialylated host cell receptors (27). To assess the receptor-binding avidity of the 2014–2016 virus population, we identified amino acid substitutions located within the RBS (Figure 2). The only amino acid variation within the RBS of the 2014–2016 viruses was residue A/T180; thus, we generated 3 RG viruses with the HA genes of wild-type SKP-827/16, which naturally contains T180, and viruses A/chicken/LH-55/2014 (LH-55/14) and A/chicken/SKP-989/2015 (SKP-989/15), which contain A180. All 3 viruses have L216 and I217. We used biolayer interferometry (31) to characterize the receptor-binding profiles of these viruses and compared them with those of UDL-01/08. LH-55/14 and SKP-989/15, with A180, had UDL-01/08-like receptor-binding profiles, and SKP-827/16, with T180, showed increased receptor-binding avidity for all tested receptor analogs, including an increase in human-receptor binding (Figure 3).
We then used biolayer interferometry on our previously generated UDL-01/08 and SKP-827/16 RG viruses containing A/T/V180 substitutions to define the effect of residue 180. Within the UDL-01/08 backbone, which naturally has A180, A180T caused an ≈50-fold increased binding avidity for 3SLN(6Su) and a >10-fold increased binding to 6SLN. A180V caused an ≈1,000-fold increased binding of 3SLN(6Su) and a >20-fold increased binding of 6SLN (Figure 4, panels A, B). Within the SKP-827/16 backbone, which naturally has T180, T180A caused an ≈30-fold decrease in binding to 3SLN(6Su) and a complete loss of quantifiable binding to 6SLN and 3SLN; T180V caused an ≈130-fold increase in binding to 3SLN(6Su), an ≈4-fold increase in binding to 6SLN, and detectable binding to 3SLN (Figure 4, panels C, D). These results indicated that A180T/V increases the receptor-binding avidity toward all sialylated receptor analogs tested and suggest that the receptor-binding phenotype of wild-type UDL-01/08 could become SKP-827/16-like through substituting A180, and vice versa.
Requirement of Enhanced Sialidase Activity by Viruses with Enhanced Binding Avidity for Elution from Erythrocytes
For new virions to be released from the surface of infected cells, the NA glycoprotein must have sufficient sialidase activity to outcompete the receptor-binding ability of its cognate HA; this equilibrium is known as HA–NA balance. We investigated A/T180A/T/V variants for their ability to elute from erythrocytes of different species. We used an adapted erythrocyte elution assay whereby different concentrations of bacterial receptor-destroying enzyme (RDE) were added to virus-hemagglutinated erythrocytes and elution monitored for 24 h at 37°C. This assay showed that viruses containing A180 eluted most rapidly, at earlier time points and lower concentrations of RDE than T/V180 viruses, indicating lower receptor avidity (Figure 5). Elution of V180 variants from chicken erythrocytes did not occur even at the highest tested RDE concentration after 24 h of incubation (Figure 5, panel B). Elution from guinea pig erythrocytes was minimal for T180 and V180 variants for which detectable elution only occurred after 24 h at the highest RDE concentrations (Figure 5, panel C). After 24 h incubation, elution from canine erythrocytes was complete for 5 of the test viruses for at least the top 4 RDE concentrations, but the SKP-827/16 T180 variant did not elute even at the highest concentration of RDE (Figure 5, panel A). In agreement with results of receptor-binding assays, we found that viruses with T/V180 have higher avidity toward sialylated receptors that naturally occur on erythrocytes and therefore require more time or better matched sialidase activity to elute efficiently. A control containing virus-hemagglutinated erythrocytes with no RDE treatment was included to show that, by 24 h, viruses containing A180 eluted from guinea pig and chicken erythrocytes but not from canine erythrocytes.
Effects of Receptor-Binding Avidity on Replication In Vitro
To evaluate further the effect on virus replication of the A/T/V180 viruses and variants, we assessed the propagation for each of our viruses by using mammalian and avian cell cultures: MDCKs, MDCK-SIAT1, and primary chicken kidney cells. These cell types express a mixture of α2,3- and α2,6-linked SA, making them suitable for assessing the correlation between growth kinetics and receptor binding (45). Viruses with HA T180 or V180 that showed greater receptor-binding avidities replicated to lower titers than did viruses with HA A180 for all cell types (Figure 6). However, at later time points in MDCK-SIAT1 cells, UDL-01/08 A180V replicated to comparable titers with UDL-01/08 A180. These differences in replication efficiency were reflected in MDCK plaque morphology: HA T180 or V180 produced smaller plaque sizes than viruses with HA A180 (Figure 7). This morphology could be seen for the UDL-01/08 and the SKP-827/16 HA backbones in a manner consistent with other avidity increasing amino acid substitutions in H9 virus subtype HAs (46).
To effectively control enzootic avian influenza virus infections, it is vital to perform poultry surveillance and virus characterization to inform vaccine composition and use and warn of potential pathogenic or zoonotic threats. The data generated in our study showed that only H9N2 avian influenza viruses were detected among poultry in Pakistan and, although genetically diverse, these viruses were distinct from H9N2 viruses in neighboring countries, implying in situ evolution rather than continuous cross-border spread. Furthermore, H9N2 avian influenza viruses from Pakistan retain internal gene cassettes that are partially derived from an H7N3 virus from Pakistan. Of particular concern is the detection of genotype PK3, which includes viruses that naturally have T180 in HA1, a dibasic motif at the HA cleavage site and truncated polymerase basic 1–F2 (Appendix). We have shown that the A180T substitution causes these viruses to be antigenically distinct from other contemporary H9N2 viruses in Pakistan, possibly leading to escape from vaccine-induced immunity. Moreover, T180 facilitates virus binding to human-like receptors, thus highlighting enhanced zoonotic potential.
Protective immunity generated by infection or conventional inactivated influenza vaccines is mediated though neutralizing antibodies against the major influenza antigen HA. However, viruses are able to evade neutralization by these antibodies through a process known as antigenic drift in human influenza viruses, whereby certain amino acid substitutions in HA are selected that directly prevent antibody binding (27,42,47). Although the mechanism of antigenic drift in avian influenza virus is poorly understood, there is evidence of comparable drivers of antigenic drift, whereby amino acid substitutions around the HA RBS decrease HI antibody titers (48). An alternative antibody escape mechanism proposed to be used by influenza viruses is to increase receptor-binding avidity, enabling HA–SA interactions to outcompete inhibition by neutralizing antibodies (49,50). In this study, we describe avian influenza A(H9N2) viruses that exhibit characteristics of adsorptive mutants in the field, and recently circulating viruses isolated from the field with HA amino acid substitutions that enhance binding avidity toward several different receptor analogs and have reduced HI and MNT titers compared with older viruses, thereby linking receptor-binding avidity to antigenicity. We also showed that HA A180T/V substitutions provide H9N2 viruses with enhanced binding to a human-like 6SLN receptor analog, although these viruses preferentially bind the avian-like 3SLN(6Su) receptor analog. This enhanced binding to 6SLN could enhance zoonotic potential of avian influenza viruses and further highlights the role of HA residue 180 as a marker for mammalian adaptation.
The ability to escape neutralization through enhanced receptor-binding avidity might come at a fitness cost. Virus replication of high-avidity viruses and mutants was attenuated in tested cell lines. An explanation for this finding might be an imbalance in the receptor-binding/cleaving synergy between virus HA and NA; additional HA or NA mutations might be required to rescue the attenuation observed in HA T180 and V180 viruses. Previous studies have investigated the role of residue V180 in mammalian adaptation of H9N2 avian influenza viruses (20,35,36). Teng et al. showed that viruses containing V180 had enhanced replication in mouse lungs and preferential receptor-binding toward α2,6-linked SA compared with α2,3-linked SA, highlighting their inherent ability to bind human-like receptors, which could be possibly attributed to possession of HA L216 (35). However, the 3SLN(6Su) receptor analog was not tested by Teng et al., and it is likely that their viruses would have binding preference for this avian-like receptor (17). Each of the viruses examined contained a 3-aa deletion in the stalk region of the NA glycoprotein, similar to that previously shown in H9N2 viruses with enhanced NA activity (25). All experiments conducted in our study used full-length wild-type UDL-01/08 NA (no stalk deletion), which is naturally paired with UDL-01/08 HA containing A180 and might not be well matched with viruses containing HA T/V180.
Yang et al. (36) showed that passage of an avian influenza A(H9N2) virus in differentiated swine airway epithelial cells led to an HA A180V substitution that facilitated α2,6-linked SA binding compare with that of parental virus, which preferentially bound α2,3-linked SA. However, these viruses did not show enhanced replication in porcine cell culture and did not have stalk deletions within the NA glycoprotein (36). Chan et al. showed attenuated growth in human lung organ culture of a human H9N2 virus containing HA V180 than in isolates containing D/E180 (20). These viruses also did not have any NA stalk deletions. Thus, we postulate that the ability of an H9N2 avian influenza virus containing HA T/V180 to cause efficient infection is at least partially dependent on a stalk deletion within its cognate NA glycoprotein, thus making HA residue 180 a good molecular marker for mammalian adaptation when considered alongside an appropriate NA. We aligned NA glycoproteins from viruses generated in this study and from aforementioned studies, but found that no additional modifications could be correlated with A/T/V180.
In conclusion, we have assessed the pathogenic and zoonotic risks posed by enzootic influenza A(H9N2) viruses in Pakistan by characterizing field isolates. Because of circulation of viruses with potential to escape vaccine-induced immunity, regular updating of vaccines to match circulating strains and protect poultry is needed in Pakistan. Furthermore, isolation of viruses from the G1 lineage with enhanced human receptor-binding avidity warrants continued surveillance for poultry and persons working with or near poultry.
Dr. Sealy is a doctoral student at The Pirbright Institute, Pirbright, UK. His research interests include evolution of influenza viruses and the effects that molecular changes have on virus fitness and the propensity to persist and spread.
Acknowledgments
We thank the veterinary staff and students at the University of Veterinary and Animal Sciences in Lahore, Pakistan, for their sample collection from poultry farms in Pakistan and Steve Martin for allowing us to use his Octet analysis software.
This study was supported by Biotechnology and Biological Sciences Research Council (BBSRC) Zoonoses and Emerging Livestock Systems (ZELS) Zoonoses and Emerging Livestock Systems (ZELS) award no. BB/L018853/1, BBSRC ZELS-AS award no. BB/N503563/1, BBSRC award no. BBS/E/I/0000198, BBSRC ISPG award no. BBS/E/I/00007034, BBS/E/I/00007035, BBS/E/I/00007038, and BBS/E/I/00007039; Cancer Research UK (FC001030); the Medical Research Council (FC001030); and the Wellcome Trust (FC001030).
References
- Dalby AR, Iqbal M. A global phylogenetic analysis in order to determine the host species and geography dependent features present in the evolution of avian H9N2 influenza hemagglutinin. PeerJ. 2014;2:e655. DOIPubMedGoogle Scholar
- Chen B, Zhang Z, Chen W. The study of avian influenza: I. The isolation and preliminary serological identification of avian influenza virus in chicken [in Chinese]. Chin J Vet Med. 1994;20:3–5.
- Zecchin B, Minoungou G, Fusaro A, Moctar S, Ouedraogo-Kaboré A, Schivo A, et al. Influenza A (H9N2) virus, Burkina Faso. Emerg Infect Dis. 2017;23:2118–9. DOIPubMedGoogle Scholar
- Aamir UB, Wernery U, Ilyushina N, Webster RG. Characterization of avian H9N2 influenza viruses from United Arab Emirates 2000 to 2003. Virology. 2007;361:45–55. DOIPubMedGoogle Scholar
- Alexander DJ. Report on avian influenza in the Eastern Hemisphere during 1997-2002. Avian Dis. 2003;47(Suppl):792–7. DOIPubMedGoogle Scholar
- Kim JA, Cho SH, Kim HS, Seo SH. H9N2 influenza viruses isolated from poultry in Korean live bird markets continuously evolve and cause the severe clinical signs in layers. Vet Microbiol. 2006;118:169–76. DOIPubMedGoogle Scholar
- Lindh E, Ek-Kommonen C, Väänänen V-M, Vaheri A, Vapalahti O, Huovilainen A. Molecular epidemiology of H9N2 influenza viruses in Northern Europe. Vet Microbiol. 2014;172:548–54. DOIPubMedGoogle Scholar
- Monne I, Hussein HA, Fusaro A, Valastro V, Hamoud MM, Khalefa RA, et al. H9N2 influenza A virus circulates in H5N1 endemically infected poultry population in Egypt. Influenza Other Respi Viruses. 2013;7:240–3. DOIPubMedGoogle Scholar
- Naeem K, Siddique N, Ayaz M, Jalalee MA. Avian influenza in Pakistan: outbreaks of low- and high-pathogenicity avian influenza in Pakistan during 2003-2006. Avian Dis. 2007;51(Suppl):189–93. DOIPubMedGoogle Scholar
- Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol. 2005;43:5760–7. DOIPubMedGoogle Scholar
- Huang Y, Li X, Zhang H, Chen B, Jiang Y, Yang L, et al. Human infection with an avian influenza A (H9N2) virus in the middle region of China. J Med Virol. 2015;87:1641–8. DOIPubMedGoogle Scholar
- Khan SU, Anderson BD, Heil GL, Liang S, Gray GC. A systematic review and meta-analysis of the seroprevalence of influenza A (H9N2) infection among humans. J Infect Dis. 2015;212:562–9. DOIPubMedGoogle Scholar
- Pawar SD, Tandale BV, Raut CG, Parkhi SS, Barde TD, Gurav YK, et al. Avian influenza H9N2 seroprevalence among poultry workers in Pune, India, 2010. PLoS One. 2012;7:e36374. DOIPubMedGoogle Scholar
- Hoa LNM, Tuan NA, My PH, Huong TTK, Chi NTY, Hau Thu TT, et al.; The Vizions Consortium. Assessing evidence for avian-to-human transmission of influenza A/H9N2 virus in rural farming communities in northern Vietnam. J Gen Virol. 2017;98:2011–6. DOIPubMedGoogle Scholar
- Thuy DM, Peacock TP, Bich VTN, Fabrizio T, Hoang DN, Tho ND, et al. Prevalence and diversity of H9N2 avian influenza in chickens of Northern Vietnam, 2014. Infect Genet Evol. 2016;44(suppl C):530–40. DOIPubMedGoogle Scholar
- Iqbal M, Yaqub T, Reddy K, McCauley JW. Novel genotypes of H9N2 influenza A viruses isolated from poultry in Pakistan containing NS genes similar to highly pathogenic H7N3 and H5N1 viruses. PLoS One. 2009;4:e5788. DOIPubMedGoogle Scholar
- Peacock TP, Benton DJ, Sadeyen J-R, Chang P, Sealy JE, Bryant JE, et al. Variability in H9N2 haemagglutinin receptor-binding preference and the pH of fusion. Emerg Microbes Infect. 2017;6:e11. DOIPubMedGoogle Scholar
- Taubenberger JK, Reid AH, Lourens RM, Wang R, Jin G, Fanning TG. Characterization of the 1918 influenza virus polymerase genes. Nature. 2005;437:889–93. DOIPubMedGoogle Scholar
- Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–8. DOIPubMedGoogle Scholar
- Chan RWY, Chan LLY, Mok CKP, Lai J, Tao KP, Obadan A, et al. Replication of H9 influenza viruses in the human ex vivo respiratory tract, and the influence of neuraminidase on virus release. Sci Rep. 2017;7:6208. DOIPubMedGoogle Scholar
- Seo SH, Hoffmann E, Webster RG. Lethal H5N1 influenza viruses escape host anti-viral cytokine responses. Nat Med. 2002;8:950–4. DOIPubMedGoogle Scholar
- Cui L, Liu D, Shi W, Pan J, Qi X, Li X, et al. Dynamic reassortments and genetic heterogeneity of the human-infecting influenza A (H7N9) virus. Nat Commun. 2014;5:3142. DOIPubMedGoogle Scholar
- Cameron KR, Gregory V, Banks J, Brown IH, Alexander DJ, Hay AJ, et al. H9N2 subtype influenza A viruses in poultry in pakistan are closely related to the H9N2 viruses responsible for human infection in Hong Kong. Virology. 2000;278:36–41. DOIPubMedGoogle Scholar
- Mitnaul LJ, Matrosovich MN, Castrucci MR, Tuzikov AB, Bovin NV, Kobasa D, et al. Balanced hemagglutinin and neuraminidase activities are critical for efficient replication of influenza A virus. J Virol. 2000;74:6015–20. DOIPubMedGoogle Scholar
- Sun Y, Tan Y, Wei K, Sun H, Shi Y, Pu J, et al. Amino acid 316 of hemagglutinin and the neuraminidase stalk length influence virulence of H9N2 influenza virus in chickens and mice. J Virol. 2013;87:2963–8. DOIPubMedGoogle Scholar
- Baigent SJ, McCauley JW. Influenza type A in humans, mammals and birds: determinants of virus virulence, host-range and interspecies transmission. BioEssays. 2003;25:657–71. DOIPubMedGoogle Scholar
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. DOIPubMedGoogle Scholar
- Shinya K, Ebina M, Yamada S, Ono M, Kasai N, Kawaoka Y. Avian flu: influenza virus receptors in the human airway. Nature. 2006;440:435–6. DOIPubMedGoogle Scholar
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. DOIPubMedGoogle Scholar
- França M, Stallknecht DE, Howerth EW. Expression and distribution of sialic acid influenza virus receptors in wild birds. Avian Pathol. 2013;42:60–71. DOIPubMedGoogle Scholar
- Lin YP, Xiong X, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, et al. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci U S A. 2012;109:21474–9. DOIPubMedGoogle Scholar
- Wan H, Perez DR. Amino acid 226 in the hemagglutinin of H9N2 influenza viruses determines cell tropism and replication in human airway epithelial cells. J Virol. 2007;81:5181–91. DOIPubMedGoogle Scholar
- Sang X, Wang A, Ding J, Kong H, Gao X, Li L, et al. Adaptation of H9N2 AIV in guinea pigs enables efficient transmission by direct contact and inefficient transmission by respiratory droplets. Sci Rep. 2015;5:15928. DOIPubMedGoogle Scholar
- Gambaryan AS, Tuzikov AB, Pazynina GV, Desheva JA, Bovin NV, Matrosovich MN, et al. 6-sulfo sialyl Lewis X is the common receptor determinant recognized by H5, H6, H7 and H9 influenza viruses of terrestrial poultry. Virol J. 2008;5:85. DOIPubMedGoogle Scholar
- Teng Q, Xu D, Shen W, Liu Q, Rong G, Li X, et al. A single mutation at position 190 in hemagglutinin enhances binding affinity for human type sialic acid receptor and replication of H9N2 avian influenza virus in mice. J Virol. 2016;90:9806–25. DOIPubMedGoogle Scholar
- Yang W, Punyadarsaniya D, Lambertz RL, Lee DC, Liang CH, Höper D, et al. Mutations during the adaptation of H9N2 avian influenza virus to the respiratory epithelium of pigs enhance sialic acid binding activity and virulence in mice. J Virol. 2017;91:e02125–16. DOIPubMedGoogle Scholar
- Naeem K, Ullah A, Manvell RJ, Alexander DJ. Avian influenza A subtype H9N2 in poultry in Pakistan. Vet Rec. 1999;145:560. DOIPubMedGoogle Scholar
- Shanmuganatham K, Feeroz MM, Jones-Engel L, Walker D, Alam S, Hasan M, et al. Genesis of avian influenza H9N2 in Bangladesh. Emerg Microbes Infect. 2014;3:e88. DOIPubMedGoogle Scholar
- Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, et al. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc Natl Acad Sci U S A. 2015;112:548–53. DOIPubMedGoogle Scholar
- Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell. 1998;95:409–17. DOIPubMedGoogle Scholar
- Baron J, Tarnow C, Mayoli-Nüssle D, Schilling E, Meyer D, Hammami M, et al. Matriptase, HAT, and TMPRSS2 activate the hemagglutinin of H9N2 influenza A viruses. J Virol. 2013;87:1811–20. DOIPubMedGoogle Scholar
- Peacock T, Reddy K, James J, Adamiak B, Barclay W, Shelton H, et al. Antigenic mapping of an H9N2 avian influenza virus reveals two discrete antigenic sites and a novel mechanism of immune escape. Sci Rep. 2016;6:18745. DOIPubMedGoogle Scholar
- Matrosovich M, Matrosovich T, Carr J, Roberts NA, Klenk H-D. Overexpression of the α-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol. 2003;77:8418–25. DOIPubMedGoogle Scholar
- Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365–94. DOIPubMedGoogle Scholar
- Ha Y, Stevens DJ, Skehel JJ, Wiley DC. X-ray structures of H5 avian and H9 swine influenza virus hemagglutinins bound to avian and human receptor analogs. Proc Natl Acad Sci U S A. 2001;98:11181–6. DOIPubMedGoogle Scholar
- Peacock TP, Benton DJ, James J, Sadeyen J-R, Chang P, Sealy JE, et al. Immune escape variants of H9N2 influenza viruses containing deletions at the haemagglutinin receptor binding site retain fitness in vivo and display enhanced zoonotic characteristics. J Virol. 2017;91:e002187–17. DOIPubMedGoogle Scholar
- Both GW, Sleigh MJ, Cox NJ, Kendal AP. Antigenic drift in influenza virus H3 hemagglutinin from 1968 to 1980: multiple evolutionary pathways and sequential amino acid changes at key antigenic sites. J Virol. 1983;48:52–60.PubMedGoogle Scholar
- Cattoli G, Milani A, Temperton N, Zecchin B, Buratin A, Molesti E, et al. Antigenic drift in H5N1 avian influenza virus in poultry is driven by mutations in major antigenic sites of the hemagglutinin molecule analogous to those for human influenza virus. J Virol. 2011;85:8718–24. DOIPubMedGoogle Scholar
- Hensley SE, Das SR, Bailey AL, Schmidt LM, Hickman HD, Jayaraman A, et al. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science. 2009;326:734–6. DOIPubMedGoogle Scholar
- Li Y, Bostick DL, Sullivan CB, Myers JL, Griesemer SB, Stgeorge K, et al. Single hemagglutinin mutations that alter both antigenicity and receptor binding avidity influence influenza virus antigenic clustering. J Virol. 2013;87:9904–10. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleOriginal Publication Date: November 14, 2018
1Current affiliation: Imperial College London, London, UK.
2Current affiliation: Chinese Academy of Agricultural Sciences, Beijing, China.
Table of Contents – Volume 25, Number 1—January 2019
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Munir Iqbal, Department of Avian Influenza, The Pirbright Institute, Ash Rd, Pirbright GU24 0NF, UK; or
Top