Volume 6, Number 6—December 2000
Research
Hantavirus Pulmonary Syndrome Associated with Monongahela Virus, Pennsylvania
Figure 2
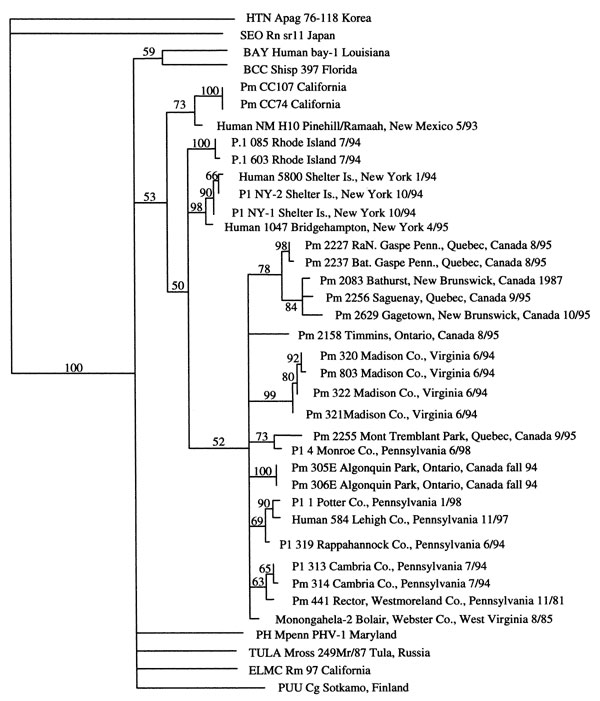
Figure 2. Phylogram of the 139-bp fragments of the G2 coding region.1 The number above the branch is the bootstrap probability (shown as percentages). HTN Apag, Hantaan virus strain 76-118, Apodemus agrarius; SEO Rn sr11, Seoul virus strain SR-11, Rattus norvegicus; BAY, Bayou virus; BCC Shisp, Black creek canal virus, Sigmodon hispidus; Pm, Peromyscus maniculatus; NM, New Mexico; Pl, Peromyscus leucopus; PH Mpenn, Prospect Hill, Microtus pennsylvanicus; TULA Mross, Tula, Microtus rossiaemeridionalis; ELMC Rm 97, El Moro Canyon virus, Reithrodontomys megalotis; PUU Cg, Puumala virus, Clethrionomys glareolus.
1Three fragments of the virus genome were amplified by nested polymerase chain reaction to allow generation of nucleotide (nt) sequences 393 nt in length for an S segment region encoding the N protein, 259 nt in length for an M segment region (M-G1) encoding the G1 protein, and 205 nt in length for an M segment region (M-G2) encoding the G2 protein (12). Sequence data were analyzed by using the Wisconsin GCG Package (version 9.1) software. Initial multiple sequence alignment was done with the GCG Pileup program, and phylogenetic analysis was performed by using the Phylogenetic Analysis Using Parsimony (and other methods) program (version 4.0) (20).