Volume 7, Number 3—June 2001
Dispatch
Expanding Drug Resistance through Integron Acquisition by IncFI Plasmids of Salmonella enterica Typhimurium
Figure 2
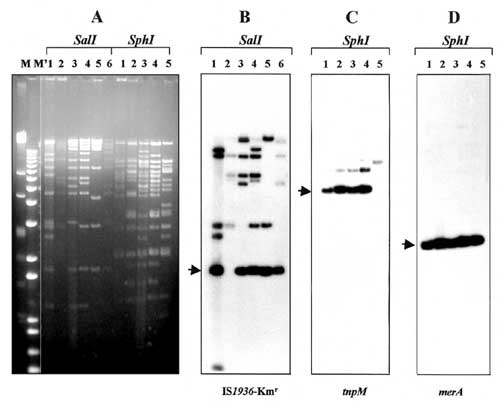
Figure 2. . Restriction enzyme analysis and detection of common genetic elements in IncFI plasmids. Numbers above each lane indicate plasmid reference numbers as defined (Table). M and M' are HindIII-digested lambda DNA and 1-kb Plus DNA ladder (GibcoBRL), respectively. A: agarose (0.8%) gel electrophoresis in 1x Tris-borate-EDTA buffer of plasmids digested with restriction endonucleases SalI and SphI. DNA was stained with ethidium bromide and visualized under UV light. B: Southern blot hybridization of SalI-digested plasmids with the IS1936-Kmr probe purified as 1.8-kb SalI fragment from plasmid pACYC184::Tn1935 (10). C and D: Southern blot hybridizations of SphI-digested plasmids with the tnpM and merA probes, respectively. The tnpM probe was amplified with primers corresponding to nt 3689-3709 and nt 4070-4050 of the released Tn21 sequence (GenEMBL accession no. AF071413). For synthesis of the merA probe, primers annealing to nt 17224-17249 and nt 17931-17907 of the Tn21 sequence were used. Plasmid pACYC184::Tn21 (10) was used as the template. Probes were labeled with [α-32P]dCTP by using a random priming kit (GibcoBRL, Rockville, MD). The arrows point to predicted hybridization fragments.
References
- Glynn MK, Bopp C, Dewitt W, Dabney P, Mokhtar M, Angulo FJ. Emergence of multidrug-resistant Salmonella enterica serotype Typhimurium DT104 infections in the United States. N Engl J Med. 1998;338:1333–8. DOIPubMedGoogle Scholar
- Prager R, Liesegang A, Rabsch W, Gericke B, Thiel W, Voigt W, Clonal relationship of Salmonella enterica serovar Typhimurium phage type DT104 in Germany and Austria. Zentralbl Bakteriol. 1999;289:399–414. DOIPubMedGoogle Scholar
- Anderson ES, Threlfall EJ, Carr JM, McConnell MM, Smith HR. Clonal distribution of resistance plasmid-carrying Salmonella typhimurium, mainly in the Middle East. J Hyg (Lond). 1977;79:425–48. DOIPubMedGoogle Scholar
- Domart A, Robineau M, Stroh A, Dubertret LM, Modai J. Septicemia due to Salmonella wien. Diagnostic, therapeutic and epidemiologic problems. Ann Med Interne (Paris). 1974;125:915–8.PubMedGoogle Scholar
- Casalino MA, Comanducci A, Nicoletti M, Maimone F. Stability of plasmid content in Salmonella wien in late phases of the epidemic history. Antimicrob Agents Chemother. 1984;25:499–501.PubMedGoogle Scholar
- Frost JA, Rowe B, Ward LR, Threlfall EJ. Characterization of resistance plasmids and carried phages in an epidemic clone of multi-resistant Salmonella typhimurium in India. J Hyg (Lond). 1982;88:193–204. DOIPubMedGoogle Scholar
- Tosini F, Visca P, Luzzi I, Dionisi AM, Pezzella C, Petrucca A, Class 1 integron-borne multiple-antibiotic resistance carried by IncFI and IncL/M plasmids in Salmonella enterica serotype Typhimurium. Antimicrob Agents Chemother. 1998;42:3053–8.PubMedGoogle Scholar
- Hall RM, Collis CM. Mobile gene cassettes and integrons: capture and spread of genes by site-specific recombination. Mol Microbiol. 1995;15:593–600. DOIPubMedGoogle Scholar
- Liebert CA, Hall RM, Summers AO. Transposon Tn21, a flagship of the floating genome. Microbiol Mol Biol Rev. 1999;63:507–22.PubMedGoogle Scholar
- Colonna B, Bernardini M, Micheli G, Maimone F, Nicoletti M, Casalino M. The Salmonella wien virulence plasmid pZM3 carries Tn1935, a multiresistance transposon containing a composite IS1936-kanamycin resistance element. Plasmid. 1988;20:221–31. DOIPubMedGoogle Scholar
- Maimone F, Colonna B, Bazzicalupo P, Oliva B, Nicoletti M, Casalino MA. Plasmids and transposable elements in Salmonella wien. J Bacteriol. 1979;139:369–75.PubMedGoogle Scholar
- National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial disc susceptibility tests. M2A2. Villanova, Pa: The Committee; 1992.
- Kado CI, Liu S. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–73.PubMedGoogle Scholar