Volume 8, Number 3—March 2002
Research
Molecular Classification of Enteroviruses Not Identified by Neutralization Tests
Figure 1
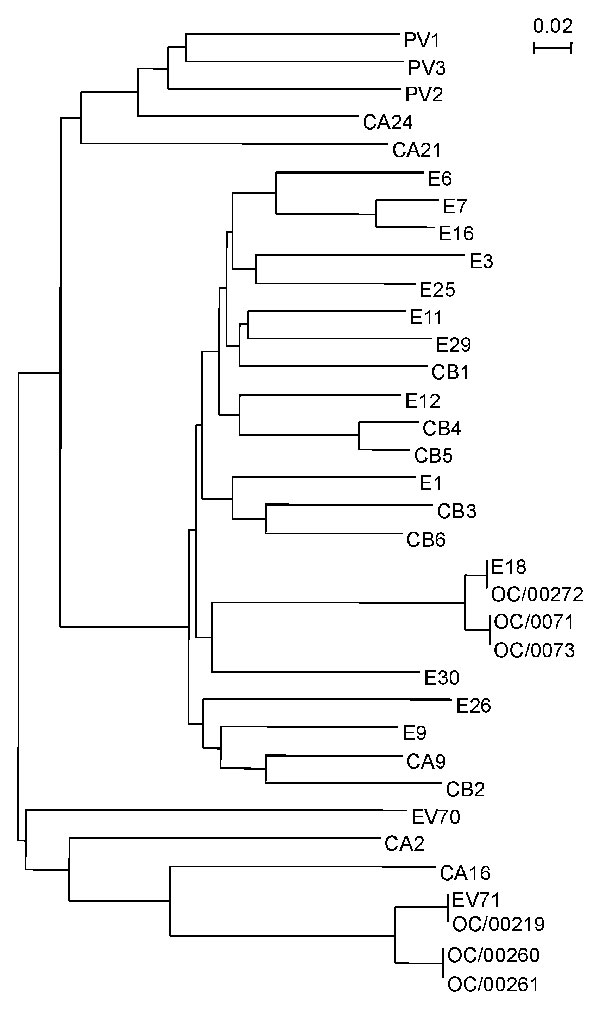
Figure 1. Phylogenetic analysis based on the human enterovirus (HEV) VP4 nucleotide sequences. The phylogenetic tree was constructed by the neighbor-joining method as implemented in CLUSTAL X program (version 1.63b). The marker denotes a measurement of the relative phylogenetic distance. The VP4 sequences of eight HEV serotypes described below are available from GenBank. The strain name and accession number are shown in parentheses: HEV1 (Bryson, AF250874), HEV12 (Travis, NC 001810), echovirus 26 (EV26, Coronel, AF117697), EV29 (JV-10, AF117698), coxsackie virus A2 (CAV2; Epsom/14448/99, AJ2296215), CAV21 (Coe, NC 001428), CAV24 (EH24/70, D90457), and HEV70 (J670/71, D00820).
Page created: July 15, 2010
Page updated: July 15, 2010
Page reviewed: July 15, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.