Volume 9, Number 2—February 2003
Research
Molecular Typing of IberoAmerican Cryptococcus neoformans Isolates
Figure 2
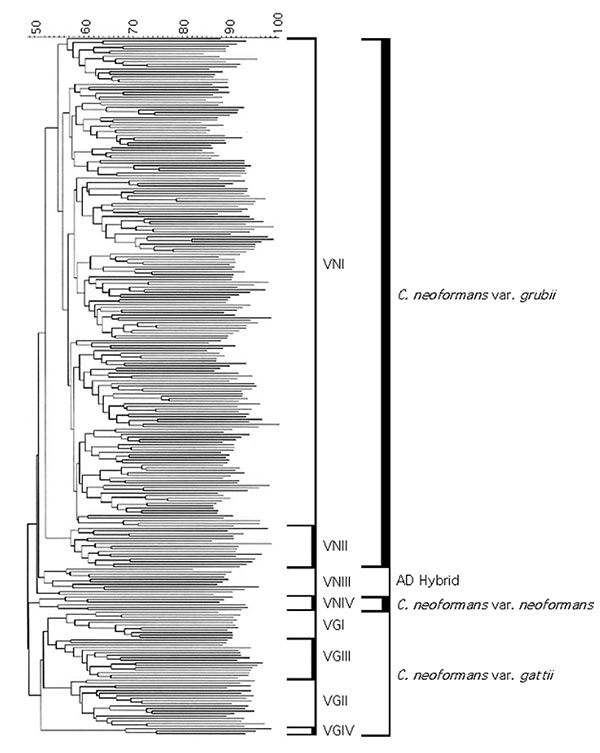
Figure 2. Dendrogram of the polymerase chain reaction-fingerprinting patterns obtained with the primer M13 from a selection of the IberoAmerican isolates studied. All the isolates fall into eight major molecular types, which fall into three major groups corresponding to Cryptococcus neoformans var. grubii, serotype A, with two molecular types VNI and VNII; C. neoformans var. neoformans, serotype D, with the molecular type VNIV; and C. neoformans var. gattii serotypes B and C, with the molecular types VGI, VGII, VGIII and VGIV. In addition to the three major clusters we can see the intermediate molecular type VNIII, representing the AD hybrids.
1Members of the IberoAmerican Cryptococcal Study Group: Argentina: Alicia Arechavala, Hospital de Infecciosas Francisco J. Muñiz, Buenos Aires; Graciela Davel, Laura Rodero, Diego Perrotta, Departamento Micología, Instituto Nacional de Enfermedades Infecciosas “Dr. Carlos Gregorio Malbrán,” Buenos Aires; Brazil: Marcia Lazera, Ricardo Pereira-Igreja, Bodo Wanke, Laboratorio de Micologia Medica, Hospital Evandro Chagas, Fundaςao Oswaldo Cruz, Rio de Janeiro; Maria Jose Mendes-Giannini, Faculdade de Ciências Farmacêuticas, Universidade Estadual Paulista (UNESP), Araraquara; Marcia S.C. Melhem, Adolfo Lutz Institute Seςao de Micologia, São Paulo; Marlene Henning-Vainstein, Centro de Biotecnologia (UFRGS), Porto Alegre; Chile: Maria Cristina Díaz, Programa de Microbiología y Micología, Universidad de Chile, Santiago; Colombia: Angela Restrepo, Corporación para Investigaciones Biológicas, Medellín; Sandra Huérfano, Instituto Nacional de Salud, Bogotá; Guatemala: Blanca Samayoa, Hospital San Juan de Dios, Guatemala City; Heidi Logeman, Universidad de San Carlos, Guatemala City; Mexico: Rubén López Martínez, Laura Rocío Castañon Olivares, Departamento de Microbiología y Parasitología, Facultad de Medicina, Universidad Nacional Autónoma de México, México City; Cudberto Contreras-Peres, José Francisco Valenzuela Tovar, Instituto Nacional de Diagnóstico y Referencia Epidemiológicos , Mexico City; Peru: Beatriz Bustamante, Instituto de Medicina Tropical Alexander Humboldt, Lima; Spain: Joseph Torres-Rodriquez, Yolanda Morera, Grup de recerca en Micologia Experimental i Clínica, Institut Municipal d’Investigació Médica, Univesitat Autonoma de Barcelona, Barcelona; Venezuela: Belinda Calvo, Universidad del Zulia, Maracaibo