Virulent Strain of Hepatitis E Virus Genotype 3, Japan
Kazuaki Takahashi

, Hiroaki Okamoto, Natsumi Abe, Manri Kawakami, Hiroyuki Matsuda, Satoshi Mochida, Hiroshi Sakugawa, Yoshiki Suginoshita, Seishiro Watanabe, Kazuhide Yamamoto, Yuzo Miyakawa, and Shunji Mishiro
Author affiliations: Toshiba General Hospital, Tokyo, Japan (K. Takahashi, N. Abe, S. Mishiro); Jichi Medical University School of Medicine, Tochigi, Japan (H. Okamoto); Kurashiki Medical Center, Okayama, Japan (M. Kawakami); Matsuda Naika Clinic, Tottori, Japan (H. Matsuda); Saitama Medical University, Saitama, Japan (S. Mochida); Heart-Life Hospital, Okinawa, Japan (H. Sakugawa); Kyoto University Graduate School of Medicine, Kyoto, Japan (Y. Suginoshita); Kagawa Prefectural Central Hospital, Kagawa, Japan (S. Watanabe); Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama (K. Yamamoto); Miyakawa Memorial Research Foundation, Tokyo (Y. Miyakawa)
Main Article
Figure 2
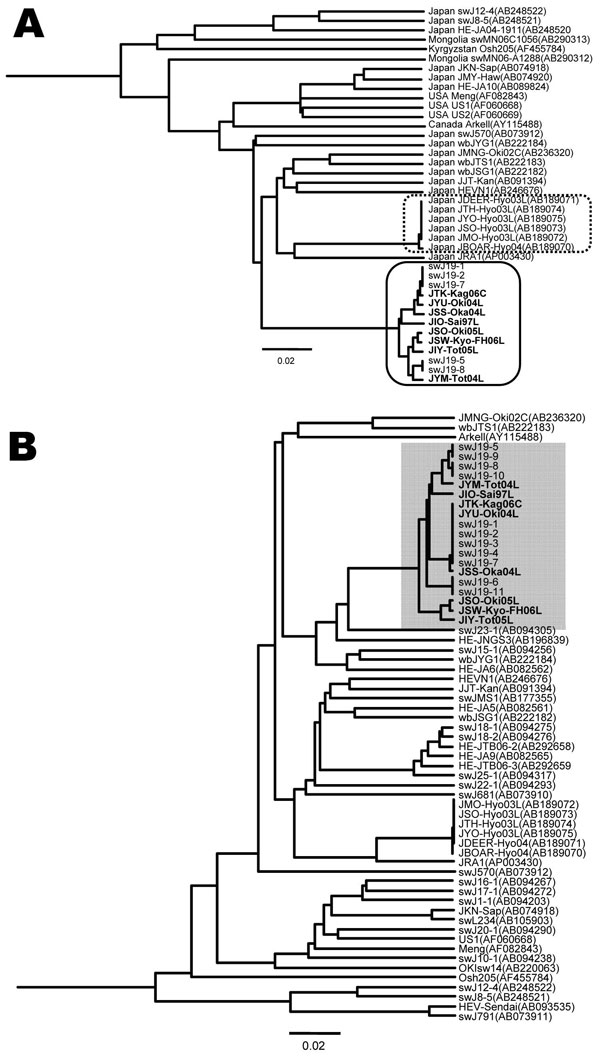
Figure 2. A) Phylogenetic tree (unweighted pair-grouping method with arithmetic means) constructed on the complete or near-complete nucleotide sequences of hepatitis E virus (HEV) genotype 3 isolates. Clustering of nucleotide sequences of 8 human patients infected with JIO strain of HEV and 5 swine infected with swJ19 strain of HEV is boxed by a solid line. Another clustering of local genotype 3 isolates from Hyogo Prefecture, Japan, is boxed by a dotted line. B) Phylogenetic tree (unweighted pair-grouping method with arithmetic means) constructed on a partial sequence of 412 nt in open reading frame (ORF) 2 (nt 5994-6405 of the US2 genome) of HEV genotype 3. Partial nucleotide sequences of 8 human JIO and 11 swine HEV swJI9 isolates (accession nos. AB094279–AB094289) are shown (shading). Analyses of full sequences of 5 of these 11 swine isolates were performed (swJ19-1, swJ19-2, swJ19-5, swJ19-7, and swJ19-8). Scale bars indicate nucleotide substitutions per site; boldface indicates isolates from humans.
Main Article
Page created: December 16, 2010
Page updated: December 16, 2010
Page reviewed: December 16, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
