Volume 27, Number 12—December 2021
Research Letter
Postmortem Stability of SARS-CoV-2 in Mouse Lung Tissue
Abstract
The infectivity of severe acute respiratory syndrome coronavirus 2 in deceased persons and organisms remains unclear. We studied transgenic K18 hACE2 mice to determine the kinetics of virus infectivity after host death. Five days after death, virus infectivity in the lung declined by >96% and RNA copies declined by 48.2%.
The safe handling and disposal of bodies of persons who have died of coronavirus disease (COVID-19) is vital for infection control. Although cremation or burial practices are mainly dictated by religious and societal customs, deaths associated with contagious illness warrant appropriate precautions. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of COVID-19, is rapidly inactivated (>2 log10) within hours on nonporous surfaces (1). In addition, several studies have detected viral RNA by reverse transcription PCR (RT-PCR) of nasopharyngeal and pharyngeal mucosal swab specimens, skin swab specimens, and tissue samples collected during autopsies at different times after death (2–5). Furthermore, infectious virus was isolated in 2 of 4 cases at 4–17 days postmortem; however, this study did not quantify virus titers to determine the loss of virus infectivity (6). A separate study found that infectious virus was undetectable after exhumation at 3–4 months postmortem (7). Overall, RNA detection by RT-PCR might not directly correlate with virus infectivity or duration of symptomatic disease.
Transgenic K18-hACE2 mice provide a surrogate model to study the kinetics of SARS-CoV-2 viral replication during infection (8) and after host death. In humans and K18-hACE2 mice, little evidence exists for extrapulmonary dissemination of SARS-CoV-2, except for neurotropism in younger mice, a finding that has not been demonstrated reliably in humans. We investigated the temporal decay of infectious SARS-CoV-2 in postmortem tissues of infected K18-hACE2 mice. All experimental procedures were conducted in accordance with the standards and approved by the Committee on the Use of Live Animals in Teaching and Research (approval no. 5511-20) at The University of Hong Kong (Hong Kong, China).
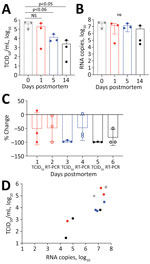
We infected twelve 14–20-week-old mice with 1 × 104 50% tissue culture infectious dose (TCID50)/25μL SARS-CoV-2 by the intranasal route. Five days later, after the mice had lost 18.8% (SD 7.77%) of their body weight, we euthanized them by ketamine/xylazil anesthesia. We wrapped each carcass in a sealable plastic bag, similar to the storage of human corpses, and stored them intact at 4°C, which is standard mortuary temperature. On days 0, 1, 5, and 14 after death, we dissected 3 carcasses and tested the lung tissue for coronavirus nucleoprotein (N) by histologic and immunohistochemistry assays (9) (Appendix Figure, panels A–H). We quantified infectious virus by culture (Figure, panel A) and viral RNA by RT-PCR (Figure, panel B) (Appendix).
Viral decay, measured using TCID50 for infectious virus and RNA copies of the N gene detected by RT-PCR, occurred over a 14-day period (Table). At day 1 we observed a 50% reduction of infectious virus and 48.8% loss of viral RNA (Figure, panels A, B). By day 5, levels of infectious virus had fallen by 96.5%, whereas viral RNA remained at 48.2% compared with day 0 (Figure, panels C, D). At day 14 only 0.7% of the initial infectious virus and 17% of viral RNA remained. Plenzig et al. (7) detected viral RNA in 2 exhumed corpses at 3 months postmortem, despite an absence of infectious virus. We used hematoxylin and eosin staining to detect viral nucleoprotein in lung tissue. We observed persistent antigen staining until day 5; by day 14, only 1 of 3 samples had detectable staining (Appendix Figure).
We euthanized the mice 5 days after infection, when the lungs had a high viral load. However, COVID-19 deaths usually occur during later stages of disease, by which time infectious viral load has decreased from the peak usually seen early during the symptomatic phase of the illness (10). We detected virus antigen in the lungs of all mice at 5 days postmortem; infectious virus had declined by 96.48%, but viral RNA declined by only 48.21%. Our results shows that infectious virus declines earlier than viral RNA or antigen in postmortem tissues.
These findings have implications for the safe handling of deceased COVID-19 patients. Infectious virus can persist on inanimate surfaces for up to 14 days at lower temperatures (<4°C), but rapidly decays in postmortem tissue samples. We observed a 96.5% decrease in infectious virus by day 5 and a 99.3% decrease by day 14. Most published postmortem studies in humans have reported viral load at the time of death using cycle threshold values rather than N gene copies as we have done; results range from 17–36 for cycle threshold values and 0–5.49 log10 for N gene copies (11). Therefore, the maximum potential risk of transmission from an infected corpse is during the first 24 hours after death. By day 5, the amount of infectious virus has decreased by 96.48%. If proper biosafety precautions and personal protective equipment are used to handle the corpse during autopsy or preparation for burial or cremation, we believe that the burial or cremation process is unlikely to spread disease.
Dr. Valkenburg is a viral immunologist at the HKU-Pasteur Research Pole, University of Hong Kong, Hong Kong, China. Her research interests include immune correlates for influenza and severe acute respiratory syndrome coronavirus 2.
Acknowledgment
This study was partly supported by the Health and Medical Research Fund (grant no. HMRF COVID-190115 to M.P. and S.A.V.), and Commissioned Research on Control of Infectious Diseases (phase III and IV) from the Health and Medical Research Fund (M.P.).
References
- Chin AWH, Chu JTS, Perera MRA, Hui KPY, Yen HL, Chan MCW, et al. Stability of SARS-CoV-2 in different environmental conditions. Lancet Microbe. 2020;1:
e10 . DOIPubMedGoogle Scholar - Heinrich F, Meißner K, Langenwalder F, Püschel K, Nörz D, Hoffmann A, et al. Postmortem stability of SARS-CoV-2 in nasopharyngeal mucosa. Emerg Infect Dis. 2021;27:329–31. DOIPubMedGoogle Scholar
- Skok K, Stelzl E, Trauner M, Kessler HH, Lax SF. Post-mortem viral dynamics and tropism in COVID-19 patients in correlation with organ damage. Virchows Arch. 2021;478:343–53. DOIPubMedGoogle Scholar
- Sablone S, Solarino B, Ferorelli D, Benevento M, Chironna M, Loconsole D, et al. Post-mortem persistence of SARS-CoV-2: a preliminary study. Forensic Sci Med Pathol. 2021;17:403–10. DOIPubMedGoogle Scholar
- Schröder AS, Edler C, Ondruschka B, Püschel K, Schädler J, Heinemann A, et al. The handling of SARS-CoV-2 associated deaths - infectivity of the body. Forensic Sci Med Pathol. 2021;17:411–8. DOIPubMedGoogle Scholar
- Plenzig S, Bojkova D, Held H, Berger A, Holz F, Cinatl J, et al. Infectivity of deceased COVID-19 patients. Int J Legal Med. 2021;135:2055–60. DOIPubMedGoogle Scholar
- Plenzig S, Holz F, Bojkova D, Kettner M, Cinatl J, Verhoff MA, et al. Detection and infectivity of SARS-CoV-2 in exhumated corpses. Int J Legal Med. 2021; Epub ahead of print. DOIPubMedGoogle Scholar
- Zheng J, Wong LR, Li K, Verma AK, Ortiz ME, Wohlford-Lenane C, et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–7. DOIPubMedGoogle Scholar
- Perera RAPM, Tso E, Tsang OTY, Tsang DNC, Fung K, Leung YWY, et al. SARS-CoV-2 virus culture and subgenomic RNA for respiratory specimens from patients with mild coronavirus disease. Emerg Infect Dis. 2020;26:2701–4. DOIPubMedGoogle Scholar
- Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382:1177–9. DOIPubMedGoogle Scholar
- Nienhold R, Ciani Y, Koelzer VH, Tzankov A, Haslbauer JD, Menter T, et al. Two distinct immunopathological profiles in autopsy lungs of COVID-19. Nat Commun. 2020;11:5086. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleOriginal Publication Date: September 23, 2021
Table of Contents – Volume 27, Number 12—December 2021
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
John Nicholls, Block T, Queen Mary Hospital, The University of Hong Kong, Pokfulam, Hong Kong, China
Top