Plasmodium falciparum pfhrp2 and pfhrp3 Gene Deletions and Relatedness to Other Global Isolates, Djibouti, 2019–2020
Volume 28, Number 10—October 2022
Eric Rogier
1
, Jessica N. McCaffery
1, Mohamed Ali Mohamed, Camelia Herman, Doug Nace, Rachel Daniels, Naomi Lucchi, Sophie Jones, Ira Goldman, Michael Aidoo, Qin Cheng, Edie A. Kemenang, Venkatachalam Udhayakumar, and Jane Cunningham
Author affiliations: Centers for Disease Control and Prevention, Atlanta, Georgia, USA (E. Rogier, J.N. McCaffery, C. Herman, D. Nace, N. Lucchi, S. Jones, I. Goldman, M. Aidoo, V. Udhayakumar); Oak Ridge Institute for Science and Education, Oak Ridge, Tennessee, USA (J.N. McCaffery); Hôpital Général Peltier, Djibouti City, Djibouti (M.A. Mohamed); Harvard T.H. Chan School of Public Health, Boston, Massachusetts, USA (R. Daniels); Broad Institute, Cambridge, Massachusetts, USA (R. Daniels); Australian Defence Force Malaria and Infectious Disease Institute, Brisbane, Queensland, Australia (Q. Cheng); World Health Organization, Geneva, Switzerland (E.A. Kemenang, J. Cunningham)
Suggested citation for this article
Abstract
Deletions of pfhrp2 and paralogue pfhrp3 (pfhrp2/3) genes threaten Plasmodium falciparum diagnosis by rapid diagnostic test. We examined 1,002 samples from suspected malaria patients in Djibouti City, Djibouti, to investigate pfhrp2/3 deletions. We performed assays for Plasmodium antigen carriage, pfhrp2/3 genotyping, and sequencing for 7 neutral microsatellites to assess relatedness. By PCR assay, 311 (31.0%) samples tested positive for P. falciparum infection, and 296 (95.2%) were successfully genotyped; 37 (12.5%) samples were pfhrp2+/pfhrp3+, 51 (17.2%) were pfhrp2+/pfhrp3–, 5 (1.7%) were pfhrp2–/pfhrp3+, and 203 (68.6%) were pfhrp2–/pfhrp3–. Histidine-rich protein 2/3 antigen concentrations were reduced with corresponding gene deletions. Djibouti P. falciparum is closely related to Ethiopia and Eritrea parasites (pairwise GST 0.68 [Ethiopia] and 0.77 [Eritrea]). P. falciparum with deletions in pfhrp2/3 genes were highly prevalent in Djibouti City in 2019–2020; they appear to have arisen de novo within the Horn of Africa and have not been imported.
Diagnosis and appropriate case management of Plasmodium falciparum infection has greatly improved in many malaria-endemic settings through the use of rapid diagnostic tests (RDTs) that detect the histidine-rich protein 2 (HRP2) antigen (1). As the only Plasmodium species infecting humans to produce this antigen, the P. falciparum parasite expresses HRP2 in abundance and releases it into the bloodstream during blood-stage infection, making this marker a very sensitive and specific target for falciparum malaria (1,2). The pfhrp2 gene is located on chromosome 8 of the parasite genome, and a paralogous gene (pfhrp3) is located on chromosome 13. The 2 protein products share common epitopes for diagnostic antibodies, enabling the HRP3 antigen to also be detected to some extent by HRP2-based RDTs (3–6).
P. falciparum produces large quantities of these antigens during human blood-stage infection, but their biologic functions are not well elucidated, and pfhrp2-deleted and pfhrp3-deleted parasites still complete the human–mosquito lifecycle successfully (7). Reports of these gene deletions have increased over the past decade from multiple countries in Africa, South America, and Asia (https://apps.who.int/malaria/maps/threats) (8). For countries that rely on HRP2-based RDTs for diagnosis of P. falciparum infection, those reports affirm the need to monitor the performance of this tool because deleted parasites could emerge and elicit false-negative results.
P. falciparum infection represents ≈99.7% of all malaria cases in sub-Saharan Africa, and ≈300 million HRP2-based RDTs are used in this region annually (9). Studies in the east Africa countries of Eritrea (10) and Ethiopia (11,12) have found high prevalence of pfhrp2/pfhrp3 deletions, forcing changes away from HRP2-based RDTs to accurately diagnose P. falciparum infections in these countries. Furthermore, it is unknown whether these deleted genotypes are a result of importation and expansion or whether de novo deletions are arising from local P. falciparum lineages. A recent report from Djibouti, which borders both Eritrea and Ethiopia, investigated 79 P. falciparum–infected patients and found ≈80% of parasites were lacking both pfhrp2 and pfhrp3 genes (13). Triggered by health workers’ reports of false-negative RDT results, we report data from an investigation of 1,002 suspected malaria patients enrolled in Djibouti City during December 2019–March 2020. Data were generated for infection-causing Plasmodium species, pfhrp2/3 genotype, concordance with laboratory antigen detection, and relatedness to other global P. falciparum parasites.
Patient Enrollment and Ethics Statement
This activity was considered by the Ministry of Health of Djibouti, the World Health Organization (WHO) Ethical Review Committee, and Centers for Disease Control and Prevention (CDC) human subjects office as nonresearch and as public health surveillance (0900f3eb81abbef6). In December 2020, a study was initiated to investigate presence of pfhrp2/3 deletions in Djibouti because of 4 specimens that tested negative by HRP2-based RDT (CareStart Malaria Combo RDT; Access Bio, https://accessbio.net) but were positive for P. falciparum lactate dehydrogenase (pf-LDH) (Bioline Malaria Ag Pf [HRP2/pLDH] Test; Abbott, https://www.abbott.com) and also confirmed for P. falciparum infection by microscopy. During January 29–March 11, 2020, consecutive patients of varying ages experiencing symptoms of malaria who sought care at Général Peltier Hospital, Djibouti City, were tested by malaria RDT with the First Response Malaria Ag (pLDH/HRP2) Combo Card Test (Premier Medical Corporation, https://www.premiermedcorp.com) and routine venipuncture for hematologic and electrolyte profiling.
Dried Blood Spot Creation
Dried blood spots (DBS) were prepared from remaining venous blood with 50–75 µL of remnant in EDTA tubes spotted on Whatman Protein Saver cards 903 or Whatmann 3M filter paper (Cytiva Life Sciences, https://www.cytivalifesciences.com). The spots were dried for >4 hours at room temperature and then stored with desiccant in sealable bags and shipped to the CDC (Atlanta, GA, USA).
Plasmodium Antigen Detection by Laboratory Multiplex Assay
DBS processing and testing for Plasmodium antigens were performed at CDC as described previously (14,15). A 6-mm punch was taken from each DBS for elution in blocking buffer (final whole blood dilution of 1:20) for the bead-based multiplex antigen detection assay of pan-Plasmodium aldolase and lactate dehydrogenase (LDH), HRP2 (and HRP3), and P. vivax LDH (PvLDH). Assay plates were run on a MAGPIX machine (Luminex Corp., https://www.luminexcorp.com) with a target of 50 beads per region.
Plasmodium Species Identification by PCR and pfhrp2 and pfhrp3 Genotyping
We selected DBS samples positive for any Plasmodium antigens for DNA extraction (DNA Mini Kit; QIAGEN, https://www.qiagen.com) and Plasmodium species–specific photo-induced electron transfer (PET) PCR and quantification of DNA (16). Samples positive for P. falciparum DNA had nested PCR reactions for single-copy pfmsp1 and pfmsp2 genes as quality control for DNA quantity and integrity (17,18). We further assayed only those samples amplifying both control genes to determine presence or absence of pfhrp2 and pfhrp3 genes. Genotyping for pfhrp2 was performed by a single-step PCR amplifying pfhrp2 inclusive of both exons (19), and genotyping for pfhrp3 was through 2 nested PCR reactions with primers specific for exon 1–2 and exon 2 regions (17,18). All genotyping reactions were run by 2 independent operators on different days and by a third operator if amplification results were discordant.
Genetic Haplotypes through Neutral Microsatellites
To assess multiplicity of infection and relatedness of P. falciparum parasites, we selected 7 neutral microsatellite (NMS) genetic markers: TA1 on chromosome 6, poly-α on chromosome 4, PfPK2 on chromosome 12, 2490 on chromosome 10, C2M34–313 on chromosome 2, C3M69–383 on chromosome 3, and TA109 on chromosome 6 (10,20). We determined the sizes of the amplification products by capillary electrophoresis on an Applied Biosystems 3130xl Genetic Analyzer (ThermoFisher Scientific, https://www.thermofisher.com) and analyzed data by using GeneMarker version 3.0.0 (SoftGenetics, https://softgenetics.com). We considered infections monoclonal if all 7 NMS had only 1 allele call. We used NMS data from previous studies to compare Djibouti results to P. falciparum parasites from other countries (Appendix Table 1).
Statistical Analysis
We compared lognormal means for HRP2/3 antigen concentration by genotype by using the 2-tailed Student t test with equal variance. For secondary analyses, we divided antigen assay signals for the entire sample set into low and high HRP2/3 antigen levels by comparing them with levels of the pLDH and pAldolase antigens as described previously (21). In brief, if blood samples were positive for pan-Plasmodium LDH or aldolase and negative for HRP2/3 antigens or had atypically low amounts of HRP2/3 relative to the pan-Plasmodium targets, we selected them as high-priority specimens with phenotypic evidence for pfhrp2/3 gene deletions (Appendix Figure 1).
We assessed NMS data and measures of relatedness by using the PopGenReport package in R (R Foundation for Statistical Computing, https://www.r-project.org) (22). For all countries, we used monoclonal infections and polyclonal infections with distinct haplotypes or dominant haplotypes for the relatedness and principal component analysis.
During January 29–March 20, 2020, of the suspected malaria cases registered at Général Peltier Hospital, 998 DBS were collected; an additional 4 samples previously collected in December 2019 were included in the sample set because they were identified as highly suspicious of pfhrp2/3 deletions on the basis of RDT results (Figure 1). Laboratory antigen screening revealed 630 (62.9%) samples were negative for all antigens; those samples were not investigated further because there was no suspicion of Plasmodium spp. infection.
We extracted DNA from the remaining 372 (37.1%) Plasmodium antigen–positive samples for further molecular investigation. Of those 372 samples, detectable Plasmodium DNA was absent in 17 (4.6%) samples, and 44 samples (11.8%) were found to be P. vivax single-species infections; samples within these 2 groups were not considered further for pfhrp2/3 deletion reporting. Of the remaining 311 samples containing P. falciparum DNA, 15 (4.8%) did not amplify both pfmsp1 and pfmsp2 single-copy genes and were not eligible for reporting of pfhrp2/3 genotypes. Of the 296 P. falciparum infections that qualified for reporting genotyping, 37 (12.5%) were a genotype of pfhrp2+/pfhrp3+, 51 (17.2%) were pfhrp2+/pfhrp3–, 5 (1.7%) were pfhrp2–/pfhrp3+, and 203 (68.6%) were a double-deletion genotype of pfhrp2–/pfhrp3– (Figure 1). For all 296 samples of P. falciparum infections successfully genotyped, 208 (70.3%) demonstrated a P. falciparum infection with pfhrp2 deletion and 254 (85.8%) showed deletion in the pfhrp3 gene.
Using the hypothetical selection method (see Methods) based on levels of pan-Plasmodium antigens compared with HRP2/3 antigens, of the 372 antigen-positive samples, 241 (64.8%) had a complete absence of HRP2/3 assay signal, 20 (5.4%) had an atypically low amount of HRP2/3 relative to other pan-Plasmodium antigens, and 111 (29.8%) had high levels of HRP2/3 relative to other pan-Plasmodium antigens (Appendix Figure 1). Of all 241 samples negative for HRP2/3 signal, 183 (75.9%) were appropriate for pfhrp2/3 genotyping, and all demonstrated a pfhrp2 deletion. When categorized into the low HRP2/3 category, of the 18 samples that qualified for genotyping, 6 (33.3%) showed a deletion of the pfhrp2 gene. If categorized into the high HRP2/3 category, of 90 samples that qualified for genotyping, the pfhrp2 gene was amplified in most (74 [82.2%]).
We found that concentrations of the HRP2 antigen (with potential supplemented signal from HRP3) were strongly associated with the different pfhrp2/3 genotypes (Figure 2). With the exception of 1 specimen, all samples found to be positive for the pfhrp2 gene (n = 88) showed high concentrations of the HRP2/3 antigen; the pfhrp2+/pfhrp3+ genotype samples showed a log-normal mean concentration of 10,794 pg/mL, and the pfhrp2+/pfhrp3– genotype samples showed a log-normal mean concentration of 18,017 pg/mL (Figure 2, panel A). Blood samples from infections with P. falciparum parasites lacking the pfhrp2 gene showed, on average, much lower concentrations of HRP2/3 antigens: log-normal mean of 421 pg/mL in pfhrp2–/pfhrp3+ samples and log-normal mean of 2.0 pg/mL in pfhrp2–/pfhrp3– samples. Concentration was significantly lower in the blood samples from pfhrp2–/pfhrp3– parasite infections than in pfhrp2+/pfhrp3+ (p<0.001), pfhrp2+/pfhrp3– (p<0.001), and pfhrp2–/pfhrp3+ (p = 0.049) parasite infections. Of the 5 infections with pfhrp2–/pfhrp3+ parasites, 2 (40.0%) showed an absence of HRP2/3 antigen signal, compared with 185/203 (91.1%) of pfhrp2–/pfhrp3– infections. Density plots of HRP2/3 antigen concentration by infecting parasite genotype illustrate these trends by genotype category (Figure 2, panel B).
In total, 65 of the Djibouti infections were chosen for NMS sequencing: 20 pfhrp2+/pfhrp3+, 20 pfhrp2+/pfhrp3–, 5 pfhrp2–/pfhrp3+, and 20 pfhrp2–/pfhrp3–. Of those, 52/65 (80.0%) were found to be monoclonal infections, and this finding did not differ significantly by genotype: pfhrp2+/pfhrp3+ (16/20 [80.0%]), pfhrp2+/pfhrp3– (15/20 [75.0%]), pfhrp2–/pfhrp3+ (4/5 [80.0%]), pfhrp2–/pfhrp3– (17/20 [85.5%]). Three of the genotypes (all except for pfhrp2–/pfhrp3+) showed a high degree of independent clustering by principal component analysis (Figure 3). Both the Jost pairwise D and Hendricks pairwise GST measures of gene differentiation found the pfhrp2–/pfhrp3– genotype to be most related to pfhrp2–/pfhrp3+ parasites (Appendix Table 2).
Comparing relatedness of P. falciparum by country (regardless of pfhrp2/3 genotype) found distinct clustering by the area of the world in which the parasites originated (Figure 4, panel A). Both Jost pairwise D and Hendricks pairwise GST found parasites from Djibouti and other parts of Africa more related to each other than to isolates from South America or Asia (Appendix Table 3, Figure 2). When repeating the analysis with only parasites from Africa, Djibouti and Ethiopia parasites were found to be most related in comparison with P. falciparum from the other 5 African countries (Appendix Figure 3). For global isolates with pfhrp2 deletions, South America and Africa parasites were not strongly related to each other; Peru and Suriname parasites had the highest principal component 1 values as well as highest Jost pairwise D and Hendricks pairwise GST in comparison with African pfhrp2-deleted parasites (Table; Figure 4, panel B). The pfhrp2 deleted parasites from Sudan, Eritrea, Ethiopia, and Djibouti showed relatively similar principal component 1 values, and Ethiopia and Djibouti pfhrp2-deleted genotypes demonstrated the closest overall clustering. Among these deleted parasites, Djibouti P. falciparum was most closely related to Ethiopia (pairwise GST 0.68) pfhrp2-deleted P. falciparum, followed by Eritrea (0.77), Sudan (0.97), Peru (0.98), and Suriname (0.99). If assessing relatedness among parasites lacking only the pfhrp3 gene, we noted similar findings; the highest degree of relatedness was seen among the Djibouti and Ethiopia parasites (Appendix Figure 4).
Confirmatory diagnosis of P. falciparum malaria through testing for the presence of HRP2 antigen by RDT has been a substantial improvement for providing appropriate case management in many malaria-endemic countries. Discovery of pfhrp2 gene deletions in natural P. falciparum populations has led to doubts about the sustained use of this antigen for diagnostic purposes (1). To date, most P. falciparum–endemic settings that rely heavily on RDTs for routine diagnostics have been found to have populations of parasites that express high amounts of HRP2 and HRP3 antigens (8,15,18,21,23,24). The ability to evade HRP2-based diagnostics (and subsequent treatment) might lead to a selective advantage for parasites with gene deletions (25). Routine surveillance of P. falciparum–endemic populations is required to ensure that primary diagnostic tools are still accurate.
Multiple recent studies in the Horn of Africa have demonstrated a high proportion of P. falciparum in the region with pfhrp2 gene deletions and that most isolates also have pfhrp3 deletions. A 2016 Eritrea health facility survey found 62.0% of P. falciparum infections consisted of parasites with both pfhrp2 and pfhrp3 deletions, all of which were also HRP2-negative by RDT (10). Of persons enrolled in health facility surveys during 2017–2018 in northern Ethiopia, pfhrp2 deletions were responsible for between 11.5%–14.9% of false-negative HRP2-RDT results (12). A survey of 3 health facilities in Djibouti in early 2019 also found these concerning results: in a small sample set of 79 P. falciparum PCR-positive patients, 86.5% demonstrated pfhrp2/3 deletions (13).
In light of previous findings, this 2019–2020 study was designed to assess the prevalence of pfhrp2/3 deletions in Djibouti City, relatedness of Djibouti P. falciparum to other global isolates, and overall relatedness of P. falciparum within the Horn of Africa compared with other global sites. Most (63%) of the 1,002 DBS samples collected from persons with malaria-like symptoms were negative for any Plasmodium antigens and did not undergo molecular assays because Plasmodium infection was not suspected. The bead-based antigen assay has been shown to detect Plasmodium infection at approximately the same level as standard PCR assays at ≈2 parasites/uL blood (15). Among the remaining 372 Plasmodium-positive samples, only 111 (29.8%) displayed moderate to high levels of HRP2/3 antigen profile indicative of P. falciparum with functional pfhrp2 or pfhrp3 genes. By those Plasmodium antigen data alone, the pervasiveness of low or absent HRP2 levels in 70% of samples from symptomatic Plasmodium-infected persons raised suspicion of either a high frequency of non–P. falciparum infections or high prevalence of P. falciparum isolates with the pfhrp2/3 genes deleted; both scenarios were found to be true in Djibouti. Of the 355 samples with detectable Plasmodium DNA by PET-PCR, 44 (12.4%) were single-species P. vivax infections; this finding is in line with previous reports of P. vivax in Djibouti, as well as in neighboring Ethiopia and Eritrea (9,26). Most (95%) of the 311 samples with P. falciparum DNA were successfully genotyped for single-copy control genes (17), reflecting high-density infections in the symptomatic study population. Of the 296 P. falciparum infections with reportable genotyping results, only 37 (12.5%) contained wild-type parasites with both pfhrp2 and pfhrp3 genes amplified. Only pfhrp3 was deleted in 17.2% of parasites, only pfhrp2 was deleted in 1.7%, and more than two thirds of all infections (68.6%) were from P. falciparum lacking both genes. The 68.6% prevalence of double-deleted parasites in Djibouti City is lower than the 86.5% previously reported by Iriart et al. (13). This high prevalence of pfhrp2/3 dual-deleted infections, coupled with our finding that most infections are monoclonal (80%), suggests that a high percentage of P. falciparum infections in Djibouti City would not be detected by HRP2-based RDTs. This level is well beyond the 5% threshold recommended by the WHO to consider a replacement of exclusive HRP2-based diagnostics for detecting P. falciparum (27), and the findings from our study have already been shared with the Djibouti Ministry of Health and WHO regional partners.
Regarding relatedness to other global isolates, the parasites found in Djibouti (regardless of genotype) clustered closely with P. falciparum haplotypes from Africa and showed greater distance to P. falciparum from the New World and Asia. Djibouti City is a large port city located on the east central coast and is home to approximately half the country’s population. Because Djibouti City is a large center of trade and population movement, some enrolled patients might have contracted P. falciparum infection in a country other than Djibouti, but travel history for participants was unavailable for this study. However, the objective genetic data show that even if some infections were acquired outside Djibouti, they all appear to be Africa-derived from more proximal countries. High relatedness (low diversity) of P. falciparum within Djibouti has been previously observed for isolates collected throughout the country within individual surveys and without substantial differences among multiple years of collection (28), although genotyping for pfhrp2/3 deletions was not performed. The relatedness of Djibouti and Ethiopia pfhrp2-deleted parasites observed in this study was closer when compared with Eritrea or Sudan isolates and very distant from pfhrp2-deleted P. falciparum from Peru and Suriname. The same finding was noted for pfhrp3-only deleted parasites, where Djibouti and Ethiopia populations practically overlie each other. This evidence points to de novo gene deletions and expansion of these deleted P. falciparum populations in the Horn of Africa rather than importation of deleted parasites from other areas of the world. Specifically for the Horn of Africa, close background lineages by NMS data for pfhrp2- and pfhrp3-deleted parasites from Djibouti and Ethiopia points to an expansion of common gene-deleted populations that exist in these adjacent countries. Eritrea parasite lineages from both wild-type and deleted P. falciparum appear to be differentiated from the Ethiopia/Djibouti lines, suggesting separate pfhrp2/3 deletion events on unique P. falciparum strains and a more distant common ancestor.
This study and its findings are subject to limitations. Though many patients exhibiting malaria symptoms were enrolled in Djibouti City, no other areas of the country are represented by this sample set, so these conclusions and estimates could not necessarily be applied nationwide. However, Djibouti City accounts for 95% of the country’s malaria case load and Général Peltier Hospital is the largest hospital in the region, and previous reports have found P. falciparum in Djibouti to be of overall low diversity (28,29). Furthermore, the Djibouti Ministry of Health considered these results sufficiently representative to mandate a nationwide RDT policy change. Without the recent travel history of enrolled participants, we cannot state all P. falciparum parasites analyzed in this study originated in Djibouti. Quality microscopy could not be performed uniformly on these blood samples, so we were unable to obtain visual confirmation of P. falciparum. In addition, because of laboratory workflow, RDT results from enrollment could not be reliably linked with venous blood samples and therefore were not compared directly with laboratory data. However, the 4 samples collected in December 2019 that triggered this investigation demonstrated pfhrp2/3 deletions causing known false-negative results by HRP2-based RDT.
In conclusion, results from both antigen detection and pfhrp2/3 molecular genotyping provide evidence of a high prevalence of symptomatic malaria cases in Djibouti caused by P. falciparum lacking functional pfhrp2/3 genes. These findings, coupled with high occurrence of monoclonal infections and single pfhrp2-deleted infections, suggest that nearly 70% of HRP2-based RDTs would return negative results for P. falciparum infection in Djibouti, which is expected to have serious negative health impacts on the community. Djibouti P. falciparum parasites with gene deletions are most closely related to other parasites in the Horn of Africa with a recent common ancestor or routine importation from Ethiopia. Gene-deleted haplotypes show no evidence of importation from South America.
Dr. Rogier is a microbiologist in the Malaria Branch, Division of Parasitic Diseases and Malaria, Center for Global Health, Centers for Disease Control and Prevention. His laboratory develops multiplex serologic assays for the detection of antibodies and antigens against infectious diseases and works to interpret this serologic data for making assumptions about population-level epidemiology and disease transmission.
Acknowledgments
We thank the staff of Général Peltier Hospital and Ghasem Zamani, who greatly assisted with this investigation.
The Bill and Melinda Gates Foundation and Centers for Disease Control and Prevention provided funding for this study.
References
Poti KE,
Sullivan DJ,
Dondorp AM,
Woodrow CJ.
HRP2: transforming malaria diagnosis, but with caveats. Trends Parasitol.
2020;
36:
112–
26.
DOIPubMedGoogle Scholar Rogier E,
Hamre KES,
Joseph V,
Plucinski MM,
Presume J,
Romilus I,
et al. Conventional and high-sensitivity malaria rapid diagnostic test performance in 2 transmission settings: Haiti 2017. J Infect Dis.
2020;
221:
786–
95.
PubMedGoogle Scholar Lee N,
Gatton ML,
Pelecanos A,
Bubb M,
Gonzalez I,
Bell D,
et al. Identification of optimal epitopes for Plasmodium falciparum rapid diagnostic tests that target histidine-rich proteins 2 and 3. J Clin Microbiol.
2012;
50:
1397–
405.
DOIPubMedGoogle Scholar Baker J,
Ho MF,
Pelecanos A,
Gatton M,
Chen N,
Abdullah S,
et al. Global sequence variation in the histidine-rich proteins 2 and 3 of Plasmodium falciparum: implications for the performance of malaria rapid diagnostic tests. Malar J.
2010;
9:
129.
DOIPubMedGoogle Scholar World Health Organization. Malaria rapid diagnostic test performance: results of WHO product testing of malaria RDTs: round 8 (2016–2018). Geneva. Organization. 2018.
Kong A,
Wilson SA,
Ah Y,
Nace D,
Rogier E,
Aidoo M.
HRP2 and HRP3 cross-reactivity and implications for HRP2-based RDT use in regions with Plasmodium falciparum hrp2 gene deletions. Malar J.
2021;
20:
207.
DOIPubMedGoogle Scholar Gamboa D,
Ho MF,
Bendezu J,
Torres K,
Chiodini PL,
Barnwell JW,
et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS One.
2010;
5:
e8091.
DOIPubMedGoogle Scholar Thomson R,
Parr JB,
Cheng Q,
Chenet S,
Perkins M,
Cunningham J.
Prevalence of Plasmodium falciparum lacking histidine-rich proteins 2 and 3: a systematic review. Bull World Health Organ.
2020;
98:
558–
568F.
DOIPubMedGoogle Scholar World Health Organization. World malaria report 2019. Geneva: The Organization; 2019.
Berhane A,
Anderson K,
Mihreteab S,
Gresty K,
Rogier E,
Mohamed S,
et al. Major threat to malaria control programs by Plasmodium falciparum lacking histidine-rich protein 2, Eritrea. Emerg Infect Dis.
2018;
24:
462–
70.
DOIPubMedGoogle Scholar Girma S,
Cheaveau J,
Mohon AN,
Marasinghe D,
Legese R,
Balasingam N,
et al. Prevalence and epidemiological characteristics of asymptomatic malaria based on ultrasensitive diagnostics: a cross-sectional study. Clin Infect Dis.
2019;
69:
1003–
10.
DOIPubMedGoogle Scholar Feleke SM,
Reichert EN,
Mohammed H,
Brhane BG,
Mekete K,
Mamo H,
et al. Plasmodium falciparum is evolving to escape malaria rapid diagnostic tests in Ethiopia. Nat Microbiol.
2021;
6:
1289–
99.
DOIPubMedGoogle Scholar Iriart X,
Menard S,
Chauvin P,
Mohamed HS,
Charpentier E,
Mohamed MA,
et al. Misdiagnosis of imported falciparum malaria from African areas due to an increased prevalence of pfhrp2/pfhrp3 gene deletion: the Djibouti case. Emerg Microbes Infect.
2020;
9:
1984–
7.
DOIPubMedGoogle Scholar Rogier E,
Nace D,
Ljolje D,
Lucchi NW,
Udhayakumar V,
Aidoo M.
Capture and Detection of Plasmodium vivax Lactate Dehydrogenase in a Bead-Based Multiplex Immunoassay. Am J Trop Med Hyg.
2020;
102:
1064–
7.
DOIPubMedGoogle Scholar Plucinski MM,
Herman C,
Jones S,
Dimbu R,
Fortes F,
Ljolje D,
et al. Screening for Pfhrp2/3-deleted Plasmodium falciparum, non-falciparum, and low-density malaria infections by a multiplex antigen assay. J Infect Dis.
2019;
219:
437–
47.
DOIPubMedGoogle Scholar Lucchi NW,
Narayanan J,
Karell MA,
Xayavong M,
Kariuki S,
DaSilva AJ,
et al. Molecular diagnosis of malaria by photo-induced electron transfer fluorogenic primers: PET-PCR. PLoS One.
2013;
8:
e56677.
DOIPubMedGoogle Scholar Abdallah JF,
Okoth SA,
Fontecha GA,
Torres RE,
Banegas EI,
Matute ML,
et al. Prevalence of pfhrp2 and pfhrp3 gene deletions in Puerto Lempira, Honduras. Malar J.
2015;
14:
19.
DOIPubMedGoogle Scholar Bharti PK,
Chandel HS,
Ahmad A,
Krishna S,
Udhayakumar V,
Singh N.
Prevalence of pfhrp2 and/or pfhrp3 gene deletion in Plasmodium falciparum population in eight highly endemic states in India. PLoS One.
2016;
11:
e0157949.
DOIPubMedGoogle Scholar Jones S,
Subramaniam G,
Plucinski MM,
Patel D,
Padilla J,
Aidoo M,
et al. One-step PCR: A novel protocol for determination of pfhrp2 deletion status in Plasmodium falciparum. PLoS One.
2020;
15:
e0236369.
DOIPubMedGoogle Scholar Uwimana A,
Umulisa N,
Venkatesan M,
Svigel SS,
Zhou Z,
Munyaneza T,
et al. Association of Plasmodium falciparum kelch13 R561H genotypes with delayed parasite clearance in Rwanda: an open-label, single-arm, multicentre, therapeutic efficacy study. Lancet Infect Dis.
2021;
21:
1120–
8.
DOIPubMedGoogle Scholar Bakari C,
Jones S,
Subramaniam G,
Mandara CI,
Chiduo MG,
Rumisha S,
et al. Community-based surveys for Plasmodium falciparum pfhrp2 and pfhrp3 gene deletions in selected regions of mainland Tanzania. Malar J.
2020;
19:
391.
DOIPubMedGoogle Scholar Daniels RF,
Chenet S,
Rogier E,
Lucchi N,
Herman C,
Pierre B,
et al. Genetic analysis reveals unique characteristics of Plasmodium falciparum parasite populations in Haiti. Malar J.
2020;
19:
379.
DOIPubMedGoogle Scholar Parr JB,
Kieto E,
Phanzu F,
Mansiangi P,
Mwandagalirwa K,
Mvuama N,
et al. Analysis of false-negative rapid diagnostic tests for symptomatic malaria in the Democratic Republic of the Congo. Sci Rep.
2021;
11:
6495.
DOIPubMedGoogle Scholar Herman C,
Huber CS,
Jones S,
Steinhardt L,
Plucinski MM,
Lemoine JF,
et al. Multiplex malaria antigen detection by bead-based assay and molecular confirmation by PCR shows no evidence of Pfhrp2 and Pfhrp3 deletion in Haiti. Malar J.
2019;
18:
380.
DOIPubMedGoogle Scholar Gatton ML,
Dunn J,
Chaudhry A,
Ciketic S,
Cunningham J,
Cheng Q.
Implications of parasites lacking Plasmodium falciparum histidine-rich protein 2 on malaria morbidity and control when rapid diagnostic tests are used for diagnosis. J Infect Dis.
2017;
215:
1156–
66.
DOIPubMedGoogle Scholar de Santi VP,
Khaireh BA,
Chiniard T,
Pradines B,
Taudon N,
Larréché S,
et al. Role of Anopheles stephensi mosquitoes in malaria outbreak, Djibouti, 2019. Emerg Infect Dis.
2021;
27:
1697–
700.
DOIPubMedGoogle Scholar World Health Organization. Response plan to pfhrp2 gene deletions. Geneva: The Organization; 2019.
Khaireh BA,
Assefa A,
Guessod HH,
Basco LK,
Khaireh MA,
Pascual A,
et al. Population genetics analysis during the elimination process of Plasmodium falciparum in Djibouti. Malar J.
2013;
12:
201.
DOIPubMedGoogle Scholar Rogier C,
Pradines B,
Bogreau H,
Koeck JL,
Kamil MA,
Mercereau-Puijalon O.
Malaria epidemic and drug resistance, Djibouti. Emerg Infect Dis.
2005;
11:
317–
21.
DOIPubMedGoogle Scholar
Suggested citation for this article: Rogier E, McCaffery JN, Mohamed MA, Herman C, Nace D, Daniels R, et al. Plasmodium falciparum pfhrp2 and pfhrp3 gene deletions and relatedness to other global isolates, Djibouti, 2019–2020. Emerg Infect Dis. 2022 Oct [date cited]. https://doi.org/10.3201/eid2810.220695
10.3201/eid2810.220695
Figure 1
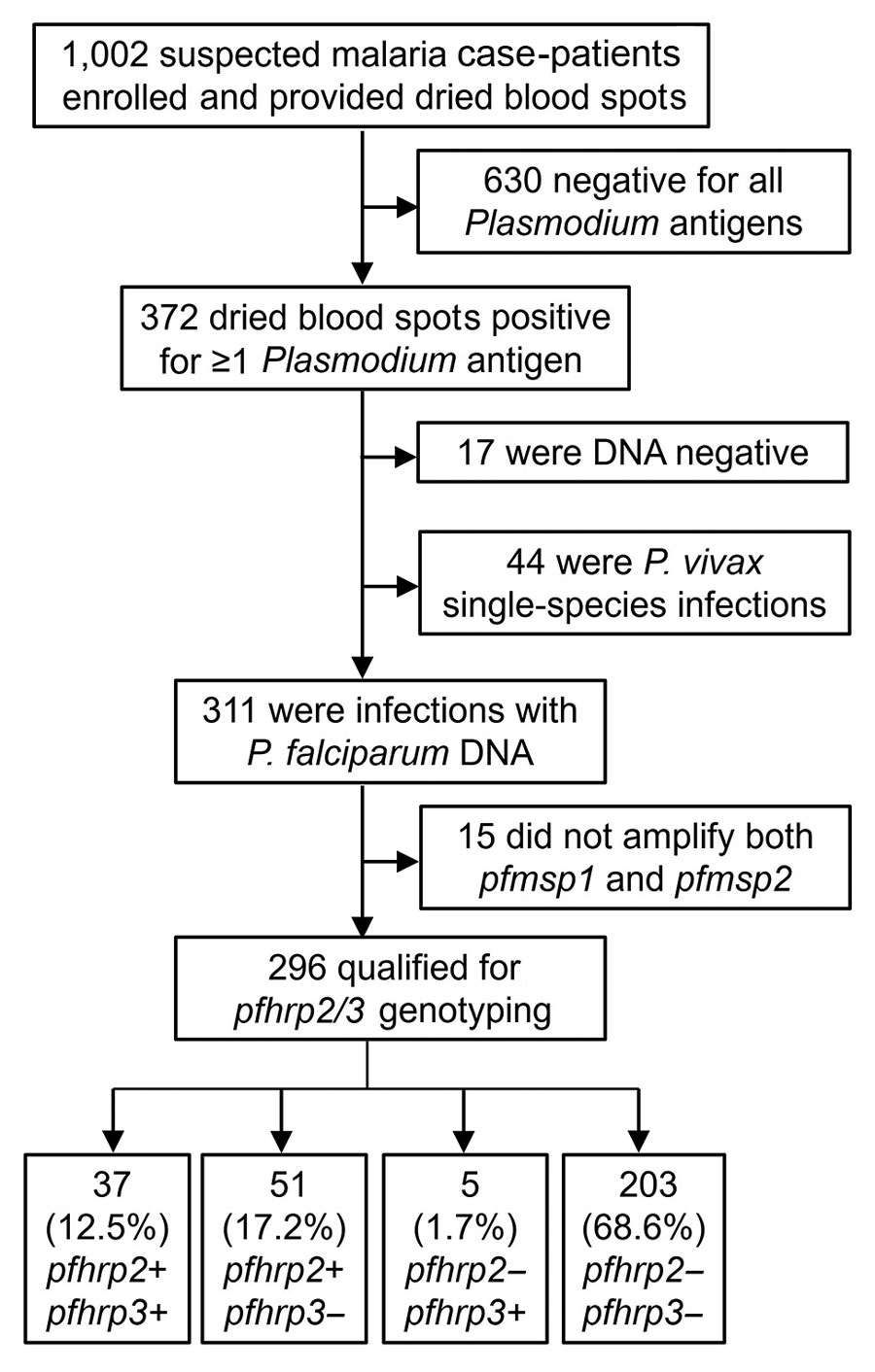
Figure 1. Flow diagram for reporting pfhrp2 and pfhrp3 genotype for all specimens in study of Plasmodium falciparum parasites with pfhrp2 and pfhrp3 deletions, Djibouti, 2019–2020. Terminal boxes display number of samples successfully genotyped for pfhrp2/3.
Figure 2
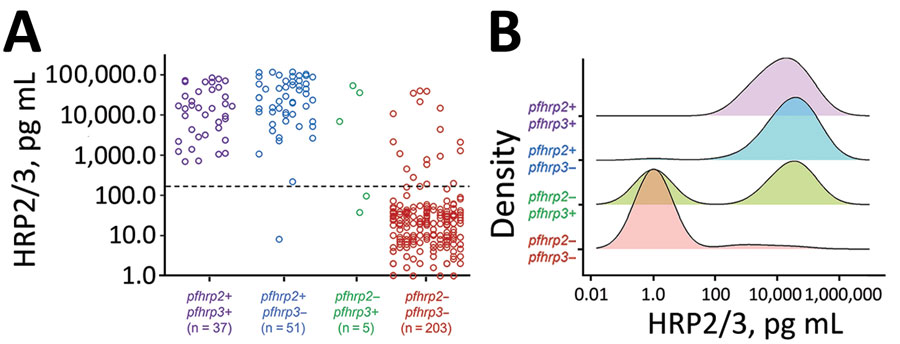
Figure 2. Distributions of HRP2/HRP3 antigen concentrations by pfhrp2 and pfhrp3 genotype for specimens in study of Plasmodium falciparum parasites with pfhrp2 and pfhrp3 deletions, Djibouti, 2019–2020. A) Individual antigen concentrations for all 296 samples successfully genotyped for pfhrp2/3. Dashed line denotes the assay level of quantitation. B) Smoothed kernel density plots for log-transformed HRP2/3 concentration by the four pfhrp2/3 genotypes. HRP2/3, histidine-rich protein 2/3.
Figure 3
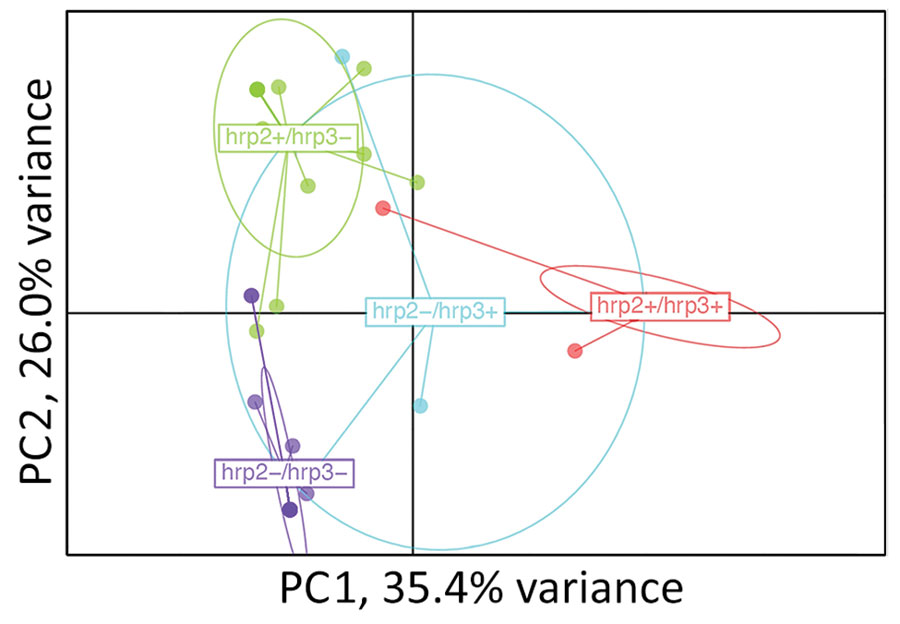
Figure 3. Relatedness of Plasmodium falciparum parasites from Djibouti, 2019–2020, with different pfhrp2 and pfhrp3 genotypes. Cluster PC analysis shown for 7 neutral microsatellite data for monogenomic infections by subpopulations: pfhrp2+/pfhrp3+ (n = 16), pfhrp2+/pfhrp3– (n = 15), pfhrp2–/pfhrp3+ (n = 4), pfhrp2–/pfhrp3– (n = 17). Plot shown with PC1 on x-axis and PC2 on y-axis with 95% confidence ellipses. PC, principal component.
Figure 4
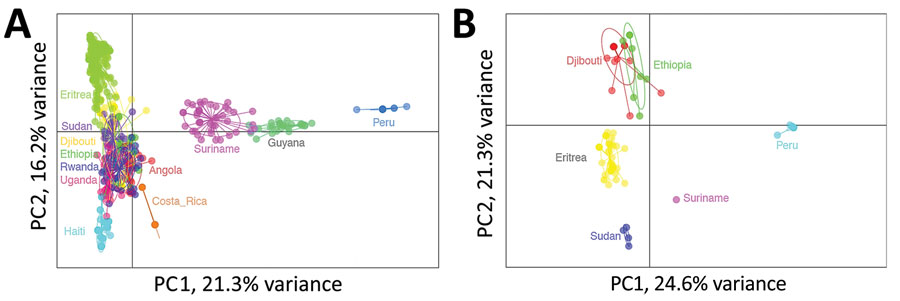
Figure 4. Relatedness of Plasmodium falciparum parasites from Djibouti, 2019–2020 with other global isolates. A) Cluster PC analysis shown for neutral microsatellite data for monogenomic infections by collection from different countries: Angola (n = 32), Costa Rica (n = 14), Djibouti (n = 52), Eritrea (n = 187), Ethiopia (n = 20), Guyana (n = 27), Haiti (n = 86), Peru (n = 18), Rwanda (n = 42), Sudan (n = 37), Suriname (n = 44), Uganda (n = 25). B) Cluster PC analysis shown for neutral microsatellite data for monogenomic infections containing pfhrp2 deletions by collection from different countries: Djibouti (n = 21), Eritrea (n = 43), Peru (n = 18), Ethiopia (n = 8), Sudan (n = 4), and Suriname (n = 1). Plots shown with PC1 on x-axis and PC2 on y-axis and 95% confidence ellipses. PC, principal component.
Table
Genetic relatedness of pfhrp2-deleted Plasmodium falciparum parasites from Djibouti, 2019–2020, compared with those from other countries*
| Comparison |
Country |
Djibouti |
Eritrea |
Ethiopia |
Peru |
Sudan |
| Jost D pairwise |
Djibouti |
|
|
|
|
|
|
Eritrea |
0.530 |
|
|
|
|
|
Ethiopia |
0.444 |
0.704 |
|
|
|
|
Peru |
0.873 |
0.888 |
0.846 |
|
|
|
Sudan |
0.920 |
0.817 |
0.879 |
0.994 |
|
|
Suriname
|
0.987
|
0.831
|
1.00
|
0.795
|
0.962
|
| Hendrick pairwise GST |
Djibouti |
|
|
|
|
|
|
Eritrea |
0.772 |
|
|
|
|
|
Ethiopia |
0.679 |
0.843 |
|
|
|
|
Peru |
0.977 |
0.972 |
0.953 |
|
|
|
Sudan |
0.972 |
0.917 |
0.941 |
0.999 |
|
|
Suriname |
0.998 |
0.949 |
1.000 |
0.991 |
0.990 |



