Multicountry Spread of Influenza A(H1N1)pdm09 Viruses with Reduced Oseltamivir Inhibition, May 2023–February 2024
Volume 30, Number 7—July 2024
Mira C. Patel
1, Ha T. Nguyen
1, Philippe Noriel Q. Pascua, Rongyuan Gao, John Steel, Rebecca J. Kondor, and Larisa V. Gubareva

Author affiliation: Centers for Disease Control and Prevention, Atlanta, Georgia, USA
Suggested citation for this article
Abstract
Since May 2023, a novel combination of neuraminidase mutations, I223V + S247N, has been detected in influenza A(H1N1)pdm09 viruses collected in countries spanning 5 continents, mostly in Europe (67/101). The viruses belong to 2 phylogenetically distinct groups and display ≈13-fold reduced inhibition by oseltamivir while retaining normal susceptibility to other antiviral drugs.
Three classes of direct-acting antivirals targeting the influenza virus matrix protein 2 (M2) ion channel, neuraminidase (NA), or polymerase cap-dependent endonuclease (CEN) are approved to treat influenza in many countries (1). Although most seasonal influenza viruses are susceptible to NA and CEN inhibitors, emergence of antiviral-resistant variants is a public health concern because of widespread resistance to M2 inhibitors and possibilities of similar resistance developing for other antiviral drugs (2). Oseltamivir, an NA inhibitor, is the drug most prescribed for influenza (2). The NA amino acid substitution H275Y, acquired spontaneously or after drug exposure, confers resistance to oseltamivir. Oseltamivir-resistant influenza A(H1N1) viruses with H275Y emerged first in Europe during 2007–2008 and rapidly spread worldwide (3). However, they were displaced by influenza A(H1N1)pdm09 (pH1N1), the swine-origin virus that caused the 2009 pandemic (4).
Monitoring oseltamivir susceptibility is a priority for the World Health Organization Global Influenza Surveillance and Response System (WHO-GISRS). In addition to H275Y, many NA substitutions in N1 subtype viruses are suspected of reducing oseltamivir susceptibility (5). Although there are no established criteria for determining clinically relevant oseltamivir resistance based on phenotypic testing, for surveillance purposes, influenza A viruses tested in NA inhibition assays are classified as displaying reduced inhibition if they have a 50% inhibitory concentration (IC50) 10-100–fold higher or as highly reduced inhibition if IC50 >100-fold higher than that of a reference (6).
The Centers for Disease Control and Prevention (CDC) monitors antiviral susceptibility of viruses submitted to the national surveillance system and those collected in other countries. Nearly all influenza-positive samples undergo next-generation sequencing. We analyzed NA sequences of submitted viruses for substitutions previously associated with reduced susceptibility (5), tested the viruses in an NA inhibition assay, and compared IC50s with a reference IC50 to determine inhibition levels (7).
During May 2023–February 2024, we analyzed 2,039 pH1N1 viruses from the United States (n = 1,274) and 38 other countries (n = 765). Four had the H275Y substitution, indicating low frequency of oseltamivir resistance. Analysis revealed NA substitution I223V in 18 and S247N in 15 viruses; those substitutions confer mildly elevated oseltamivir IC50 (<10-fold). We also detected 17 viruses carrying both substitutions, I223V + S247N (Table 1; Appendix Table 1). As expected, single mutants exhibited normal inhibition by oseltamivir and other NA inhibitors in NA inhibition assay (Table 2). The 6 viruses with I223V + S247N displayed 13- to 16-fold reduced inhibition for oseltamivir and normal inhibition (<4-fold) for other NA inhibitors (Table 2; Appendix Table 2). Both single and dual mutants remained susceptible to the CEN inhibitor baloxavir (Table 2).
The dual mutants were collected in the United States and 4 other countries during August–November 2023 (Table 1; Appendix Table 1), which prompted us to explore when dual mutants emerged and how broadly single or dual mutants were circulating worldwide. Analysis of available NA sequences from the GISAID EpiFlu database (https://www.gisaid.org) revealed that pH1N1 viruses with single mutation I223V (n = 110) or S247N (n = 203) were found in many countries (Figure 1, panel A). In addition to the 17 viruses we sequenced, 84 additional dual mutants were identified (total n = 101). Together, they were collected in 15 countries spanning 5 continents (Africa, Asia, Europe, North America, and Oceania) (Figure 1, panel B). The first dual mutant was collected from Canada in May 2023, and the latest were collected from 4 countries (France, the Netherlands, Spain, and the United Kingdom) during January–February 2024. Most dual mutants were detected in the Netherlands (n = 30), France (n = 24), Bangladesh (n = 11), Oman (n = 9), and the United Kingdom (n = 9); fewer were found in Hong Kong (n = 4), Niger (n = 3), Australia (n = 2), Spain (n = 2), and the United States (n = 2). One dual mutant each was detected in Canada, Ethiopia, Maldives, Norway, and Sweden (Figure 1, panel B; Appendix Tables 1, 3).
On the basis of NA phylogenetic analysis, we determined that most single S247N mutants belonged to either subclade C.5 (16%) or the most abundantly sequenced subclade, C.5.3 (80%) (8). All subclade C.5.3 viruses share substitution S200N located at the antibody binding family site VI (Figure 1, panel A). Most single I223V mutants (92%) also belonged to subclade C.5.3 and formed a distinct branch with substitution S366N located at the antibody binding family site III. Within that branch, 2 separate introductions of S247N have occurred, giving rise to dual I223V + S247N mutants that could be divided into distinct groups 1 and 2 (Figure 2). Only 9 dual mutants collected in 6 countries belonged to group 1 and shared an additional substitution R257K. Group 2 encompassed 92 dual mutants from countries with multiple detections (i.e., France, Netherlands, Bangladesh, the United Kingdom, Oman, and Niger) (Figure 1, panel B).
To further characterize the dual mutants, we performed hemagglutinin (HA) phylogenetic analysis. Two major HA clades, 6B.1A.5a.2a (5a.2a) and 6B.1A.5a.2a.1 (5a.2a.1), were seen globally during this period (8,9). Viruses belonging to HA subclades 5a.2a_C.1 (55%–61%) and 5a.2a.1_C.1.1.1 (13%–39%) predominated. Of note, all group 1 dual mutants had HA from 5a.2a_C.1, represented by the previous vaccine prototype virus A/Sydney/5/2021 (Figure 2). Conversely, most group 2 dual mutants had HA from 5a.2a.1_C.1.1.1, represented by the current vaccine prototype virus A/Victoria/4897/2022, and most shared 2 HA changes: R45K in HA1 and L2I in HA2 (Figure 2).
We report the emergence and intercontinental spread of pH1N1 viruses displaying reduced susceptibility to oseltamivir resulting from acquisition of NA-I223V + S247N mutations. Emergence of the dual mutants was also recently noticed by researchers in Hong Kong (10). The dual mutants that we tested retained susceptibility to other approved influenza antiviral drugs, including baloxavir. Analysis of available sequence data revealed that dual mutants have been in global circulation since May 2023; overall detection frequency was low (0.67%, 101/15,003). However, those data may not necessarily represent the actual proportion of what was in circulation because of differences in surveillance and sequencing strategies in each country.
Substitutions at residues 223 or 247 were previously reported and occurred spontaneously in circulating viruses (5,11,12). pH1N1 viruses with S247N circulated in several countries during 2009–2011 (11), and influenza B viruses with I223V (I221V in B numbering) were found in several US states during 2010–2011 (12). Isoleucine at 223 is a highly conserved framework residue in the NA active site. The S247N substitution may alter the hydrogen bonding network of the active site and the conformation of the residue E277 side chain, thereby weakening oseltamivir binding (11). Changes at 223 or 247 are monitored because they can enhance drug resistance by combining with mutations at other residues (11,13,14).
Rapid spread of dual mutants to countries on different continents suggests no substantial loss in their replicative fitness and transmissibility. I223V was shown to alter NA activity (14), and change at 247 may produce a similar effect, which warrants the question whether signature substitution(s) of NA subclade C.5.3 and of the branch to which the dual mutants belong (i.e., S200N, S366N) could serve as prerequisites for emergence of dual mutants. All group 1 dual mutants had an additional substitution R257K, which was previously associated with restoring NA activity of viruses with the H275Y substitution (15). Conversely, most group 2 viruses acquired HA from subclade 5a.2a.1_C.1.1.1 by reassortment, which may have helped to restore the functional HA and NA balance. Acquisition of the antigenically distinct HA could further enhance the spread of group 2 dual mutants. Our study highlights the need to closely monitor evolution of dual mutants because additional changes may further affect susceptibility to antiviral drugs or provide a competitive advantage over circulating wild-type viruses.
Dr. Patel and Dr. Nguyen are scientists in the Influenza Division, National Center for Immunization and Respiratory Diseases, CDC, Atlanta. Their research interests are respiratory viruses and antiviral therapeutics targeting viral or host proteins, and their research focuses on influenza viruses, molecular characterization of antiviral resistance mechanisms, and development of in vitro assays for monitoring drug susceptibility.
Acknowledgments
We are grateful to the US public health laboratories and other laboratories participating in the WHO-GISRS for productive collaboration with the CDC National Center for Immunization and Respiratory Diseases Influenza Division and their timely submission of influenza viruses. We acknowledge the efforts and contributions of WHO-GISRS laboratories and other laboratories that have performed genetic sequencing and submitted influenza sequence data to GISAID EpiFlu. We also acknowledge the valuable contributions of colleagues from the Virology, Surveillance and Diagnosis Branch of the CDC Influenza Division.
This study was supported by the CDC Influenza Division.
References
Holmes EC,
Hurt AC,
Dobbie Z,
Clinch B,
Oxford JS,
Piedra PA.
Understanding the impact of resistance to influenza antivirals. Clin Microbiol Rev.
2021;
34:
e00224–
20.
DOIPubMedGoogle Scholar Meijer A,
Lackenby A,
Hungnes O,
Lina B,
van-der-Werf S,
Schweiger B,
et al.;
European Influenza Surveillance Scheme.
Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 season. Emerg Infect Dis.
2009;
15:
552–
60.
DOIPubMedGoogle Scholar Garten RJ,
Davis CT,
Russell CA,
Shu B,
Lindstrom S,
Balish A,
et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science.
2009;
325:
197–
201.
DOIPubMedGoogle Scholar Meetings of the WHO working group on surveillance of influenza antiviral susceptibility – Geneva, November 2011 and June 2012. Wkly Epidemiol Rec.
2012;
87:
369–
74.
PubMedGoogle Scholar Okomo-Adhiambo M,
Mishin VP,
Sleeman K,
Saguar E,
Guevara H,
Reisdorf E,
et al. Standardizing the influenza neuraminidase inhibition assay among United States public health laboratories conducting virological surveillance. Antiviral Res.
2016;
128:
28–
35.
DOIPubMedGoogle Scholar Aksamentov I,
Roemer C,
Hodcroft EB,
Neher RA.
Nextclade: clade assignment, mutation calling and quality control for viral genomes. J Open Source Softw.
2021;
6:
3773.
DOIGoogle Scholar Leung RC, Ip JD, Chen LL, Chan WM, To KK. Global emergence of neuraminidase inhibitor-resistant influenza A(H1N1)pdm09 viruses with I223V and S247N mutations: implications for antiviral resistance monitoring. Lancet Microbe. 2024;S2666-5247(24)00037-5.
Hurt AC,
Lee RT,
Leang SK,
Cui L,
Deng YM,
Phuah SP,
et al. Increased detection in Australia and Singapore of a novel influenza A(H1N1)2009 variant with reduced oseltamivir and zanamivir sensitivity due to a S247N neuraminidase mutation. Euro Surveill.
2011;
16:
19884.
DOIPubMedGoogle Scholar Garg S,
Moore Z,
Lee N,
McKenna J,
Bishop A,
Fleischauer A,
et al. A cluster of patients infected with I221V influenza b virus variants with reduced oseltamivir susceptibility—North Carolina and South Carolina, 2010-2011. J Infect Dis.
2013;
207:
966–
73.
DOIPubMedGoogle Scholar Centers for Disease Control and Prevention (CDC).
Oseltamivir-resistant 2009 pandemic influenza A (H1N1) virus infection in two summer campers receiving prophylaxis—North Carolina, 2009. MMWR Morb Mortal Wkly Rep.
2009;
58:
969–
72.
PubMedGoogle Scholar Pizzorno A,
Bouhy X,
Abed Y,
Boivin G.
Generation and characterization of recombinant pandemic influenza A(H1N1) viruses resistant to neuraminidase inhibitors. J Infect Dis.
2011;
203:
25–
31.
DOIPubMedGoogle Scholar Bloom JD,
Nayak JS,
Baltimore D.
A computational-experimental approach identifies mutations that enhance surface expression of an oseltamivir-resistant influenza neuraminidase. PLoS One.
2011;
6:
e22201.
DOIPubMedGoogle Scholar
Suggested citation for this article: Patel MC, Nguyen HT, Pascua PNQ, Gao R, Steel J, Kondor RJ, et al. Multicountry spread of influenza A(H1N1)pdm09 viruses with reduced oseltamivir inhibition, May 2023–February 2024. Emerg Infect Dis. 2024 Jul [date cited]. https://doi.org/10.3201/eid3007.240480
10.3201/eid3007.240480
Figure 1
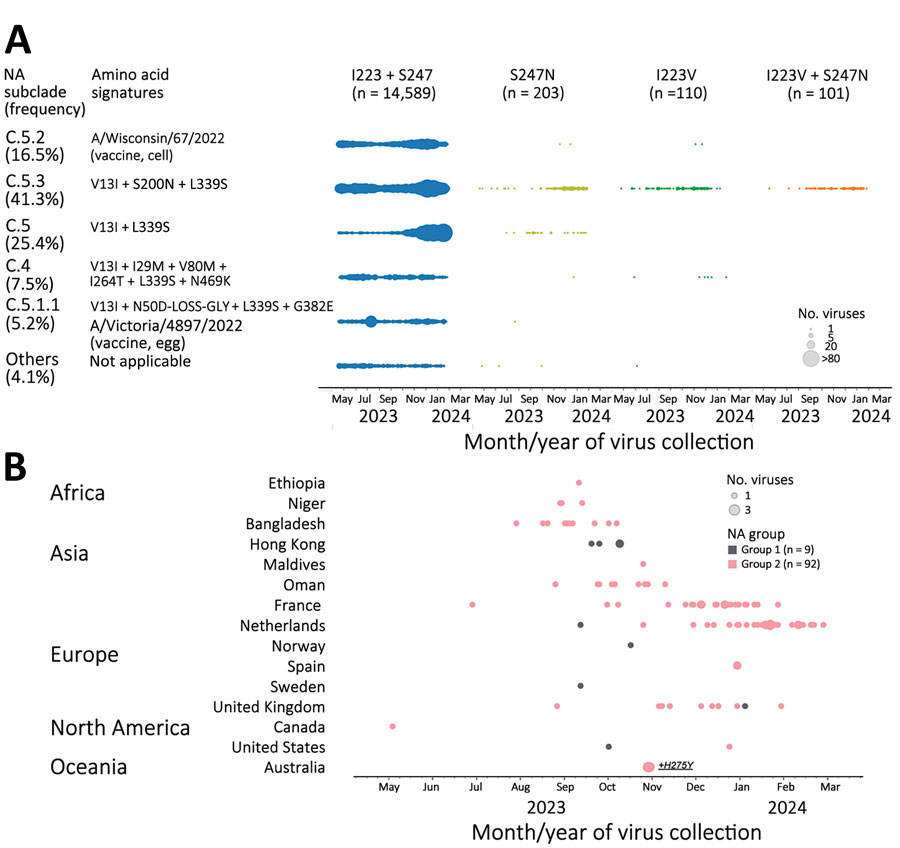
Figure 1. Detection of influenza A(H1N1)pdm09 viruses with dual NA-I223V + S247N substitutions through NA inhibitors susceptibility surveillance conducted by the Centers for Disease Control and Prevention and analysis of available sequences (GISAID EpiFlu, https://www.gisaid.org, accessed March 11, 2024), May 2023–February 2024. A total of 15,003 NA sequences of pH1N1 viruses (duplicate sequences excluded: 2,039 from Centers for Disease Control and Prevention and the remaining 12,964 from GISAID EpiFlu) were analyzed to screen for amino acid substitutions at residues 223 and 247. A) Introduction of single substitution (I223V or S247N) or dual substitutions (I223V + S247N) across NA subclades of pH1N1 viruses circulating during May 2023–February 2024. Amino acid signatures of NA subclades are shown in comparison to A/Wisconsin/67/2022, the Northern Hemisphere 2023–2024 vaccine cell prototype virus for the pH1N1 component. Vaccine viruses, A/Wisconsin/67/2022 and A/Victoria/4897/2022 (Northern Hemisphere 2023–2024 vaccine egg prototype virus), represented NA subclades C.5.2 and C.5.1.1, respectively. C.5.3 was the subclade most abundantly sequenced (41.3% frequency), followed by other minor subclades: C.5 (25.4%), C.5.2 (16.5%), C.4 (7.5%), C.5.1.1 (5.2%), and others (4.1%). Most viruses with single S247N substitution belonged to dominant NA subclade C.5.3 and minor subclade C.5. Conversely, most viruses with single I223V substitution and all viruses with dual I223V + S247N substitutions belonged to NA subclade C.5.3. B) Spatiotemporal distribution of dual mutant viruses. Dual mutants were divided into 2 groups based on their NA sequence difference. Group 1 shared additional substitution R257K not found in group 2. The small group 1 had 9 dual mutants, and the large group 2 had 92 dual mutants. The first dual mutant belonging to group 2 was collected in Canada at the end of May 2023, and most dual mutants were collected between September 2023 and February 2024. Two dual mutant viruses collected in Australia also contained NA-H275Y. NA, neuraminidase.
Figure 2
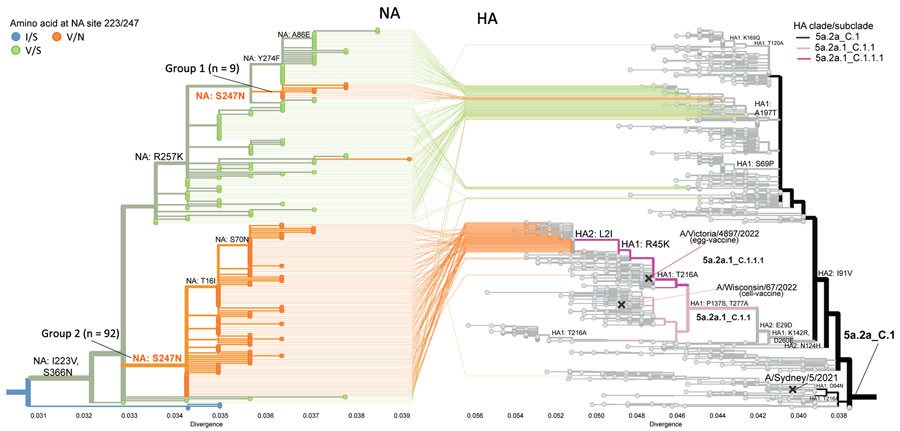
Figure 2. Tanglegram showing influenza A(H1N1)pdm09 phylogenies for NA gene (left) and HA gene (right) from susceptibility surveillance conducted by the Centers for Disease Control and Prevention and analysis of available sequences (GISAID EpiFlu, https://www.gisaid.org, accessed March 11, 2024), May 2023–February 2024. The NA-HA tangle tree was constructed by using Nextclade (8) and visualized by Auspice (https://auspice.us). NA phylogenetic tree is zoomed to show only subclade C.5.3 with 2 groups of dual I223V + S247N mutants. All dual mutant viruses shared >6 NA amino acid substitutions (V13I, S200N, I223V, S247N, L339S, S366N) compared with vaccine prototype virus A/Wisconsin/67/2022. Group 1 shared an additional substitution R257K not observed in group 2. Tree tips colored in blue indicate viruses with wild type amino acids at residues 223 and 247 (i.e., I223 and S247); green shows single I223V mutants; and orange shows dual mutants. Small group 1 with 9 dual mutants and large group 2 with 92 dual mutants are indicated. The NA sequence and corresponding HA of each virus are connected by lines. Group 1 dual mutants have HA 5a.2a_C.1. Only 2 group 2 dual mutants (A/British Columbia/PHL1108/2023 collected in May 2023 and A/France/IDF-RELAB-IPP24993/2023 collected in October 2023) have HA 5a.2a_C.1; remaining group 2 dual mutants shared HA 5a.2a.1_C.1.1.1. HA 5a.2a_C.1 is represented by A/Sydney/5/2021, the Southern Hemisphere 2023 vaccine egg/cell prototype virus. HA 5a.2a.1_C.1.1.1 is represented by A/Victoria/4897/2022, the Northern Hemisphere 2023–2024 vaccine egg prototype virus. The Northern Hemisphere 2023–2024 vaccine cell prototype virus, A/Wisconsin/67/2022, represents HA 5a.2a.1_C.1.1. HA, hemagglutinin; NA, neuraminidase.
Table 1
Influenza A(H1N1)pdm09 viruses with amino acid substitutions in neuraminidase that may affect inhibition by oseltamivir*
| NA substitution† |
No. viruses with NA substitution |
Viruses collected from countries (no. in each) |
| H275Y |
4 |
Argentina (1),‡ Panama (1), USA (2) |
| I223V |
18 |
Abu Dhabi (5), Bangladesh (5), Bahrain (2), Canada (1), Costa Rica (1), USA (4) |
| S247N |
15 |
Abu Dhabi (2), Bhutan (3), Brazil (1), Canada (1), Hong Kong (1), Oman (1), USA (6) |
| I223V + S247N§ |
17 |
Bangladesh (11), Hong Kong (1), Maldives (1), Niger (3), USA (1) |
Table 2
Antiviral susceptibility of available influenza A(H1N1)pdm09 virus isolates with NA-I223V or NA-S247N or NA-I223V + S247N substitutions*
| NA substitution† |
No. virus isolates tested |
NA inhibitors IC50, nM, average ± SD (fold)‡
|
CEN inhibitor baloxavir EC50, nM, average ± SD (fold)§ |
| Oseltamivir |
Zanamivir |
Peramivir |
Laninamivir |
| Test viruses |
|
|
|
|
|
|
| I223V |
9 |
0.71 ± 0.17 (3) |
0.23 ± 0.03 (1) |
0.08 ± 0.01 (1) |
0.25 ± 0.01 (1) |
1.15 ± 0.15 (2) |
| S247N |
6 |
0.82 ± 0.16 (4) |
0.34 ± 0.05 (2) |
0.19 ± 0.01 (3) |
0.55 ± 0.03 (3) |
1.46 ± 0.62 (2) |
I223V + S247N
|
6
|
2.71 ± 0.20 (13)
|
0.49 ±0.02 (3)
|
0.29 ± 0.01 (4)
|
0.64 ± 0.02 (3)
|
0.67 ± 0.14 (1)
|
| Reference |
|
|
|
|
|
|
| Median IC50/EC50
2022–2023 |
253 |
0.21 |
0.19 |
0.07 |
0.21 |
0.70 |

