Volume 15, Number 7—July 2009
Dispatch
Co-infections with Chikungunya Virus and Dengue Virus in Delhi, India
Abstract
Aedes aegypti mosquitoes are common vectors for dengue virus and chikungunya virus. In areas where both viruses cocirculate, they can be transmitted together. During a dengue outbreak in Delhi in 2006, 17 of 69 serum samples were positive for chikungunya virus by reverse transcription–PCR; 6 samples were positive for both viruses.
Chikungunya virus (CHIKV) was isolated in Tanganyika (now Tanzania) in 1953 (1). In Asia, this virus is transmitted almost exclusively by Aedes aegypti mosquitoes. India had its first CHIKV outbreak in 1963; it was followed by epidemics in other parts of the country (2). Recently, massive outbreaks of CHIKV have been reported from many islands in the Indian Ocean (3). Chikungunya outbreaks in India were reported in 2005, and 1.4 million chikungunya cases were reported from different states (3).
Estimated annual incidence of disease caused by dengue virus (DENV) is 50–100 million cases of dengue fever and 250,000 cases of dengue hemorrhagic fever; mortality rate is 25,000 per year in tropical and subtropical countries. Like CHIKV, DENV is also transmitted by Ae. aegypti and is endemic to urban and semiurban areas of India (4).
In Asia, the CHIKV-affected areas overlap with DENV-endemic areas (5,6) and provide opportunities for mosquitoes to become infected with both viruses. Co-infection with 2 dengue viruses (DENV-1 and DENV-4) was reported in Puerto Rico in 1982 (7). Since then, many cases of concurrent infections with multiple DENV serotypes have been reported in many countries. Since 2005, co-infections with >2 DENV serotypes have been reported in Delhi, India (8). Co-infections with DENV and CHIKV were reported in Calcutta, India, in 1967 (5). Subsequent serologic investigations in southern India indicated that the 2 viruses can coexist in the same host (9). We report detection by reverse transcription–PCR of co-infections with CHIKV and DENV in clinical samples obtained during the 2006 dengue outbreak in Delhi, India.
Acute-phase blood samples were collected from 69 patients with clinically suspected cases of DENV/CHIKV co-infection. Viral RNA was extracted from serum samples by using the MagNA Pure Compact Nucleic Acid Isolation System (Roche Diagnostics, Basel, Switzerland). Published primers and cycling conditions were used for the amplification of DENV (10) and CHIKV (11). cDNA was synthesized by using avian myeloblastosis virus reverse transcriptase (Promega Corp., Madison, WI, USA) and downstream consensus primer (D2) for DENV and random hexamers for CHIKV. DENV typing was performed by using second-round amplification with type-specific primers (10). The amplified products were visualized by electrophoresis on 2% agarose gels. Because samples were received during a dengue outbreak, ethical clearance was not required.
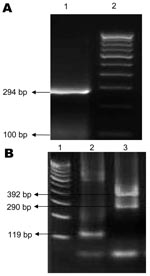
Partial nucleotide sequences of the envelope 1 (E1) gene (294 bp) of CHIKV were determined by using an automated 310 DNA sequencer (Applied Biosystems Inc., Foster City, CA, USA). Sequences were aligned, analyzed, subjected to homology search by BLAST analysis (www.ncbi.nlm.nih.gov/Education/BLASTinfo/information3.html), and submitted to GenBank (accession nos. EU727159–63 and EF539265). Phylogenetic analysis of CHIKV sequences (Table) was conducted by using ClustalW (www.ebi.ac.uk/Tools/clustalw2/index.html) and MEGA version 3.1 software (12), Kimura 2-parameter distances, and neighbor-joining algorithms.
Of 69 samples tested, DENV RNA was detected in 48 and CHIKV RNA in 17. Of the 17 CHIKV-positive samples, 6 were co-infected with DENVs. Three of the 6 samples from patients co-infected with CHIKV/DENV contained DENV-3; 1 contained DENV-4, and 2 contained 2 DENV serotypes (1 contained DENV-3 and DENV-4 and 1 contained DENV-3 and DENV-1) (Figure 1).
Of the 17 CHIKV-positive patients, 10 were male and 6 were female. Information regarding age, sex, and clinical features was not available for 1 patient. Thirteen samples were from adults (>12 years of age) and 3 were from children (<12 years of age).
Retrospective analysis of medical records identified clinical information for 6 patients co-infected with DENV and CHIKV. All 6 patients had fever, headache, joint pain, and low thrombocyte counts (<100,000/mm3). The patients with only CHIKV infection had fever, headache, and joint pain. Of the 6 patients with co-infections, 2 had dengue hemorrhagic fever with central nervous system (CNS) involvement. CNS involvement and hemorrhagic manifestations may be caused by DENVs because these manifestations are common in patients infected with DENV; CNS involvement has been documented in persons with DENV infections (13). In 2 patients with CNS involvement, 1 was infected with DENV-3 and 1 was infected with DENV-4. Of the 6 patients with co-infections, 5 fully recovered and 1 died.
Phylogenetic analysis of partial E1 gene sequences demonstrated that all CHIKV strains from Delhi grouped with isolates obtained during 2006 from southern India and islands in the Indian Ocean and belonged to the Central/East African genotype (Figure 2). This finding indicates that during 2006 similar strains were circulating throughout India. Isolates obtained in India during 1963–1973 clustered with isolates from Thailand (Thai 62–78) and formed a separate cluster in the Asian genotype.
For many years, it appeared that CHIKV had disappeared from India, but late in 2005 the virus reemerged on Reunion Island and in India (3). Confirmed cases of CHIKV infection have been reported from Delhi, Haryana, Uttar Pradesh, and Rajasthan provinces in northern India, although these states did not have large-scale epidemics (14).
DENV infections are endemic to northern India; in recent years, increasing trends of cocirculation of multiple DENV serotypes in Delhi suggest that DENVs are becoming hyperendemic to this region (8). During 2006, DENV and CHIKV were detected in Delhi (14). Because the clinical features of DENV and CHIKV are similar, CHIKV infections may go undiagnosed in DENV-endemic areas. In India, Ae. aegypti mosquitoes are primary vectors for DENV and CHIKV, and opportunities for co-infections in humans are increased by the feeding behavior of the mosquito (15), low socioeconomic conditions, and high population density.
We report co-infections with DENV and CHIKV in India after a long absence of the viruses in this region. It is difficult to comment on increased severity of illness in patients with DENV/CHIKV co-infections because the number of patients tested was small. Additional clinical information is needed to determine the influence of co-infections on clinical expression of dengue and chikungunya fever.
Our study indicates that co-infections with CHIKV and DENV occur in areas where these 2 viruses cocirculate. Concurrent infections may result in illness with overlapping signs and symptoms, making diagnosis and treatment difficult for physicians. Repeated outbreaks of dengue, recent activity of CHIKV, and CHIKV/DENV co-infections in the Delhi area suggest that the epidemiology of these viruses is changing in this region and that these viruses are becoming endemic to this region. Thus, in clinically suspected cases of dengue or chikungunya fever, it is advisable to test for both viruses in areas where they cocirculate.
Mr Chahar is a doctoral student at the All India Institute of Medical Sciences in New Delhi, India. His research interest is the development of silent RNA–based approaches for inhibition of multiplication of respiratory syncytial virus and human metapneumovirus.
Acknowledgments
We thank Ashok Saini, Salek Chand, and Devender Kumar for excellent technical support; and Chandra P. Prasad and Sailesh Bajpai for critically reading the manuscript and making valuable suggestions.
This study was supported by All India Institute of Medical Sciences and the government of India. H.S.C. was supported by a fellowship from the Indian Council of Medical Research.
References
- Ross RW. The Newala epidemic III; the virus: isolation, pathogenic properties and relationship to the epidemic. J Hyg (Lond). 1956;54:177–91. DOIPubMedGoogle Scholar
- Shah KV, Gibbs CJ Jr, Banerjee G. Virological investigation of the epidemic of haemorrhagic fever in Calcutta: isolation of three strains of chikungunya virus. Indian J Med Res. 1964;52:676–83.PubMedGoogle Scholar
- Ravi V. Re-emergence of chikungunya virus in India. Indian J Med Microbiol. 2006;24:83–4.PubMedGoogle Scholar
- Gubler DJ. Dengue. In: Monath TP, editor. The arboviruses: epidemiology and ecology. Vol. II. Boca Raton (FL): CRC Press; 1988. p. 223–260.
- Myers RM, Carey DE. Concurrent isolation from patient of two arboviruses, chikungunya and dengue type 2. Science. 1967;157:1307–8. DOIPubMedGoogle Scholar
- Mackenzie JS, Chua KB, Daniels PW, Eaton BT, Field HE, Hall RA, Emerging viral diseases of Southeast Asia and the Western Pacific. Emerg Infect Dis. 2001;7:497–504.PubMedGoogle Scholar
- Gubler DJ, Kuno G, Sather GE, Waterman SH. A case of natural concurrent human infection with two dengue viruses. Am J Trop Med Hyg. 1985;34:170–3.PubMedGoogle Scholar
- Bharaj P, Chahar HS, Pandey A, Diddi K, Dar L, Guleria R, Concurrent infections by all four dengue virus serotypes during an outbreak of dengue in 2006 in Delhi, India. Virol J. 2008;5:1. DOIPubMedGoogle Scholar
- Yergolkar PN, Tandale BV, Arankalle VA, Sathe PS, Sudeep AB, Gandhe SS, Chikungunya outbreaks caused by African genotype, India. Emerg Infect Dis. 2006;12:1580–3.PubMedGoogle Scholar
- Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase–polymerase chain reaction. J Clin Microbiol. 1992;30:545–51.PubMedGoogle Scholar
- Hasebe F, Parquet MC, Pandey BD, Mathenge EG, Morita K, Balasubramaniam V, Combined detection and genotyping of chikungunya virus by a specific reverse transcription–polymerase chain reaction. J Med Virol. 2002;67:370–4. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. DOIPubMedGoogle Scholar
- Lum LC, Lam SK, Choy YS, George R, Harun F. Dengue encephalitis: a true entity? Am J Trop Med Hyg. 1996;54:256–9.PubMedGoogle Scholar
- Directorate General of Health Services Ministry of Health and Family Welfare. National Vector Borne Disease Control Program. State-wise status of chikungunya fever in India, 2006 [cited 2008 May 10]. Available from http://www.nvbdcp.gov.in/Doc/CHK.pdf
- Scott TW, Naksathit A, Day JF, Kittayapong P, Edman JD. A fitness advantage for Aedes aegypti and the viruses it transmits when females feed only on human blood. Am J Trop Med Hyg. 1997;57:235–9.PubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 15, Number 7—July 2009
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Shobha Broor, Virology Division, Department of Microbiology, Rm 2105, All India Institute of Medical Sciences, New Delhi 110029, India
Top