Volume 15, Number 9—September 2009
Dispatch
Genetic Differences between Avian and Human Isolates of Candida dubliniensis
Abstract
When Candida dubliniensis isolates obtained from seabird excrement and from humans in Ireland were compared by using multilocs sequence typing, 13 of 14 avian isolates were genetically distinct from human isolates. The remaining avian isolate was indistinguishable from a human isolate, suggesting that transmission may occur between humans and birds.
Candida dubliniensis is an opportunistic yeast species phenotypically and genetically closely related to C. albicans, the most common cause of Candida infection. However, C. dubliniensis is less pathogenic and is most commonly associated with superficial infection in immunocompromised persons. Although C. albicans has frequently been isolated from avian and animal sources (1–4), the recent study by Nunn et al. identified C. dubliniensis from a nonhuman source (5). These isolates were obtained from the surface of Ixodes uriae ticks that lived in cracks filled with seabird excrement at 2 locations at a seabird breeding colony on Great Saltee Island off the southeastern coast of Ireland.
Multilocus sequence typing (MLST) is an informative tool for investigating the population structure and epidemiology of many bacterial and fungal species (6). We have used MLST to show that C. dubliniensis has less genetic diversity than C. albicans and that C. dubliniensis isolates comprise 3 distinct clades (C1, C2, and C3), which correspond to described internally transcribed spacer (ITS) region genotypes 1–4 (7). Two other research groups recently used MLST to show genetic differences between C. albicans isolates from humans and animals (3,4). The purpose of our study was to use MLST, the presence or absence of a previously identified polymorphism in the CDR1 gene (8), and mating type analysis to determine genetic relatedness between avian-associated and human C. dubliniensis isolates and whether avian-associated isolates are a source of human opportunistic infections.
To obtain avian-associated C. dubliniensis isolates from a novel geographic site, fresh seabird excrement was sampled from the campus of Trinity College Dublin, ≈150 km north of Great Saltee Island by using nitrogen-gassed VI-PAK sterile swabs (Sarstedt-Drinagh, Wexford, Ireland). Samples were plated within 2 h of collection on CHROMagar Candida medium (CHROMagar, Paris, France), incubated at 30°C for 48 h, and identified as described (7,9–12). Three new C. dubliniensis isolates were obtained from 134 fecal samples. Like isolates from Great Saltee Island (5), these 3 isolates obtained directly from freshly deposited herring gull (Larus argentatus) excrement were ITS genotype 1 (13). Because the isolates originally described by Nunn et al. (5) were obtained from the surface of ticks living in avian excrement, avian-associated isolates refers to avian excrement–associated isolates. The avian-associated isolates were compared with 31 human C. dubliniensis strains belonging to MLST clade C1 as previously reported (7), and 5 additional C. dubliniensis clade C1 human isolates from Ireland (Table).
Isolates were assigned a diploid sequence type (DST) on the basis of genotype numbers for the 8 loci in the recommended C. dubliniensis MLST typing scheme (7) (Table). Six new DSTs were identified in 13 of 14 avian-associated isolates because of the identification of 2 new exZWF1b alleles that were found exclusively in avian- associated isolates. DST2 was the only previously identified DST (isolate AV7, Table). DST 31 was the most frequently (5/14 isolates) found DST in avian-associated C. dubliniensis isolates, all 5 of which were from Great Saltee Island (5). Four isolates belonged to DST 27, three from Great Saltee Island and 1 from Dublin (Table).
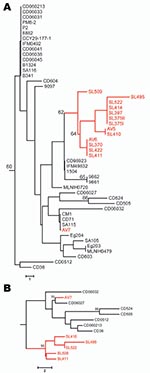
Polymorphic sites (n = 36) from the 8 MLST loci (7) of all 50 clade C1 human and avian-associated C. dubliniensis isolates were concatenated and used to construct a neighbor-joining tree (MEGA software program version 3.1 [14]), which included all known clade C1 DSTs identified. Thirteen of 14 avian-associated C. dubliniensis isolates, 11 from Great Saltee Island (5) and 2 from Dublin, formed a distinct subgroup within clade C1 (Figure, panel A). This same subgroup was also identified in trees generated by using the unweighted pair group method with arithmetic mean, maximum parsimony, and maximum likelihood, and based on related sequence types (BURST) analysis. To test for genetic separation between human and avian-associated isolates obtained from the same country, a neighbor-joining tree was constructed by using 13 avian-associated and human clade C1 isolates from Ireland, each of which represented unique DSTs. The tree showed the robustness of the avian-associated subgroup within a population of human isolates from the same country, and the distribution of avian-associated and human isolates differed significantly (p = 0.025, by Fisher exact test) (www.exactoid.com/fisher/index.php) (Figure, panel B).
The prevalence of a common point mutation, previously identified in the CDR1 gene of some ITS genotype 1 isolates, was determined for avian and human C. dubliniensis isolates as described previously (8). All 14 avian-associated isolates had the TAG polymorphism (Table) compared with 19 (53%) of 36 human clade C1 isolates. The mating types of the isolates were determined by multiplex PCR amplification by using 2 pairs of mating type locus (MTL)–specific primers. A 535-bp amplimer was generated with primers MTLa1-F (5′-TGAAAATGAAGACAATGCGA-3′) and MTLa1-R (5′-CATCTTTTTCTGCTATCAATTC-3′) in the presence of MTL type a DNA, and a 615-bp product resulted from primers MTLα1-F (5′-ATGAATTCACATCTGGAGGC-3′) and MTLα1-R (5′-CTGTTAATAGCAAAGCAGCC-3′) in the presence of MTL type α DNA. Amplification reactions contained 10 pmol of each of the forward and reverse primers, 2.5 mmol/L MgCl2, 10 mmol/L Tris-HCl, pH 9.0 at 25°C, 10 mmol/L KCl, 0.1% (vol/vol) Triton X-100, 1.25 U GoTaq polymerase (Promega, Madison, WI, USA), and 25 μL of template DNA in a total volume of 50 μL. Cycling conditions were at 94°C for 10 min; 30 cycles at 94°C for 1 min, 55°C for 2 min, and 72°C for 3 min; and a final step at 72°C for 10 min.
Of the 14 avian excrement–associated isolates, 13 were MTLa homozygous (a/a), (Table). Only 4 (11.1%) of 36 human clade C1 isolates were homozygous for MTLa; 28 (77.7%) of 36 were heterozygous for MTL (a/α). A previous study also reported that 17 (20.7%) of 82 human C. dubliniensis isolates were homozygous for MTLa (15). The gull isolate AV7 was indistinguishable from human isolates CD71, SA115, and CM1 by MLST and has the same mating type (a/α). We propose that AV7 may be a human isolate that colonized a gull scavenging on the Trinity College Dublin campus. The TAG polymorphism and mating type data from the avian-associated isolates suggest a highly clonal population.
The avian-associated C. dubliniensis isolates investigated belong to MLST clade C1, which includes most human isolates. However, most (13/14) of the avian-associated isolates form a distinct subgroup within this clade, which suggests that despite the low level of variation within C. dubliniensis, a distinct avian subpopulation may be present. This suggestion is supported by the observation that 2/3 isolates (AV5 and AV6) obtained in Dublin belonged to the same subpopulation (defined by MLST, CDR1, and MTL loci) as isolates obtained from Great Saltee Island, which is 150 km from Dublin. Similar data suggesting genetic separation and differential clade distribution between human and animal populations of C. albicans have been reported (3,4). The presence of the avian-associated subgroup within the most predominant clade (C1), which had previously only been identified in human isolates, and the close genetic relatedness between isolates, in particular gull isolate AV7, suggests that transmission between the 2 hosts can occur. However, in this instance the most likely direction of transfer is from human to bird.
Ms McManus is a doctoral candidate in the Microbiology Research Unit at Dublin Dental School and Hospital, Trinity College Dublin. Her research interests focus on analysis of the population structure of the novel yeast pathogen C. dubliniensis by using MLST.
Acknowledgments
We thank Oscar Merne for assistance with collection of avian excrement samples and identification of specific species of birds at sampling sites.
This study was supported by the Microbiology Research Unit, Dublin Dental School and Hospital.
References
- Buck JD. Isolation of Candida albicans and halophilic Vibrio spp. from aquatic birds in Connecticut and Florida. Appl Environ Microbiol. 1990;56:826–8.PubMedGoogle Scholar
- Edelmann A, Kruger M, Schmid J. Genetic relationship between human and animal isolates of Candida albicans. J Clin Microbiol. 2005;43:6164–6. DOIPubMedGoogle Scholar
- Wrobel L, Whittington JK, Pujol C, Oh SH, Ruiz MO, Pfaller MA, Molecular phylogenetic analysis of a geographically and temporally matched collection of Candida albicans isolates from humans and nonmigratory wildlife in central Illinois. Eukaryot Cell. 2008;7:1475–86. DOIPubMedGoogle Scholar
- Jacobsen MD, Bougnoux ME, d’Enfert C, Odds FC. Multilocus sequence typing of Candida albicans isolates from animals. Res Microbiol. 2008;159:436–40. DOIPubMedGoogle Scholar
- Nunn MA, Schaefer SM, Petrou MA, Brown JR. Environmental source of Candida dubliniensis. Emerg Infect Dis. 2007;13:747–50.PubMedGoogle Scholar
- Odds FC, Jacobsen MD. Multilocus sequence typing of pathogenic Candida species. Eukaryot Cell. 2008;7:1075–84. DOIPubMedGoogle Scholar
- McManus BA, Coleman DC, Moran G, Pinjon E, Diogo D, Bougnoux ME, Multilocus sequence typing reveals that the population structure of Candida dubliniensis is significantly less divergent than that of Candida albicans. J Clin Microbiol. 2008;46:652–64. DOIPubMedGoogle Scholar
- Moran G, Sullivan D, Morschhauser J, Coleman D. The Candida dubliniensis CdCDR1 gene is not essential for fluconazole resistance. Antimicrob Agents Chemother. 2002;46:2829–41. DOIPubMedGoogle Scholar
- Coleman D, Sullivan D, Harrington B, Haynes K, Henman M, Shanley D, Molecular and phenotypic analysis of Candida dubliniensis: a recently identified species linked with oral candidosis in HIV-infected and AIDS patients. Oral Dis. 1997;3(Suppl 1):S96–101.PubMedGoogle Scholar
- Al Mosaid A, Sullivan DJ, Coleman DC. Differentiation of Candida dubliniensis from Candida albicans on Pal’s agar. J Clin Microbiol. 2003;41:4787–9. DOIPubMedGoogle Scholar
- Pincus DH, Coleman DC, Pruitt WR, Padhye AA, Salkin IF, Geimer M, Rapid identification of Candida dubliniensis with commercial yeast identification systems. J Clin Microbiol. 1999;37:3533–9.PubMedGoogle Scholar
- Donnelly SM, Sullivan DJ, Shanley DB, Coleman DC. Phylogenetic analysis and rapid identification of Candida dubliniensis based on analysis of ACT1 intron and exon sequences. Microbiology. 1999;145:1871–82. DOIPubMedGoogle Scholar
- Gee SF, Joly S, Soll DR, Meis JF, Verweij PE, Polacheck I, Identification of four distinct genotypes of Candida dubliniensis and detection of microevolution in vitro and in vivo. J Clin Microbiol. 2002;40:556–74. DOIPubMedGoogle Scholar
- Kumar S, Tamura K, Nei M. MEGA3: Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5:150–63. DOIPubMedGoogle Scholar
- Pujol C, Daniels KJ, Lockhart SR, Srikantha T, Radke JB, Geiger J, The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot Cell. 2004;3:1015–27. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 15, Number 9—September 2009
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
David C. Coleman, Microbiology Research Unit, Division of Oral Biosciences, Dublin Dental School and Hospital, Trinity College Dublin, Dublin 2, Ireland;
Top