Volume 18, Number 11—November 2012
Dispatch
Fatal Respiratory Infections Associated with Rhinovirus Outbreak, Vietnam
Figure
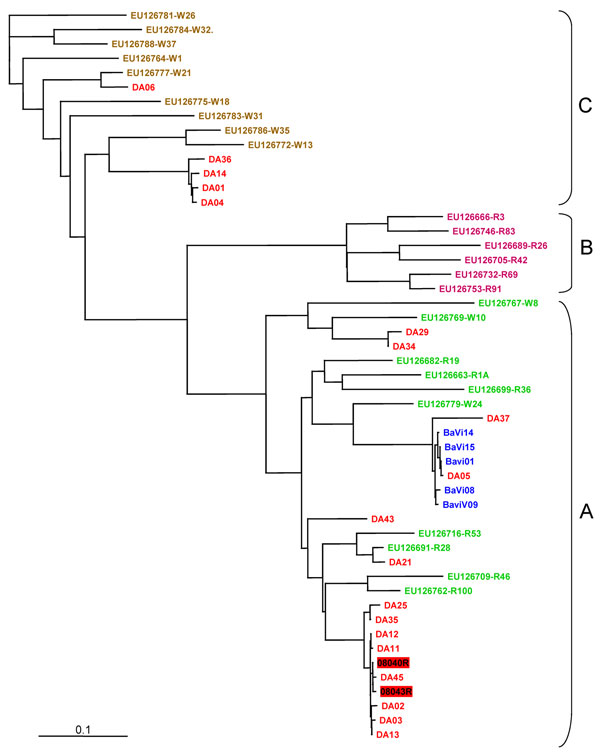
Figure. . . . Genotyping of human rhinovirus (HRV) from outbreak and control specimens by P1–P2 sequence analysis. We compared 19 HRVs from the outbreak orphanage (red), 5 HRVs from control orphanage blue), and the following P1-P2 sequences in GenBank, according to Lee et al. (3). HRV-A group (green): EU126769, EU126779, EU126767, EU126699, EU126682, EU126663, EU126716, EU126691, EU126709, EU126762; HRVB group (orange): EU126732, EU126689, EU126666, EU126753, EU126705, EU126746, HRV-C group (brown): EU126786, EU126772, EU126775, EU126777, EU126783, EU126788, EU126784, EU126781, EU126764. Two HRV sequences from hospitalized patients were included (HRV number in red box). Multiple sequence alignment was performed by using the BioEdit program package (Ibis Biosciences, Carlsbad, CA, USA). Nucleotide distances were analyzed with DNAdist, the neighbor-joining tree of BioEdit package. The consensus tree was visualized by TREEVIEW v1.6.6 (Institute of Biomedical and Life Sciences, University of Glasgow, Glasgow, UK). Scale bar indicates nucleotide substitutions per site.
References
- World Health Organization. Children: reducing mortality. World Health Organization Factsheet no. 178. 2012. [cited 2012 Aug 29]. http://www.who.int/mediacentre/factsheets/fs178/en/index.html
- Peng D, Zhao D, Liu J, Wang X, Yang K, Xicheng H, Multipathogen infections in hospitalized children with acute respiratory infections. Virol J. 2009;6:155. DOIPubMedGoogle Scholar
- Lee WM, Kiesner C, Pappas T, Lee I, Grindle K, Jartti T, A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS ONE. 2007;2:e966. DOIPubMedGoogle Scholar
- Kiang D, Yagi S, Kantardjieff KA, Kim EJ, Louie JK, Schnurr DP. Molecular characterization of a variant rhinovirus from an outbreak associated with uncommonly high mortality. J Clin Virol. 2007;38:227–37. DOIPubMedGoogle Scholar
- Kim JO, Hodinka RL. Serious respiratory illness associated with rhinovirus infection in a pediatric population. Clin Diagn Virol. 1998;10:57–65. DOIPubMedGoogle Scholar
- Briese T, Renwick N, Venter M, Jarman RG, Ghosh D, Kondgen S, Global distribution of novel rhinovirus genotype. Emerg Infect Dis. 2008;14:944–7. DOIPubMedGoogle Scholar
- Yoshida LM, Suzuki M, Yamamoto T, Nguyen HA, Nguyen CD, Nguyen AT, Viral pathogens associated with acute respiratory infections in central Vietnamese children. Pediatr Infect Dis J. 2010;29:75–7. DOIPubMedGoogle Scholar
- Savolainen C, Laine P, Mulders MN, Hovi T. Sequence analysis of human rhinoviruses in the RNA-dependent RNA polymerase coding region reveals large within-species variation. J Gen Virol. 2004;85:2271–7. DOIPubMedGoogle Scholar
- Xiang Z, Gonzalez R, Xie Z, Xiao Y, Liu J, Chen L, Human rhinovirus C infections mirror those of human rhinovirus A in children with community-acquired pneumonia. J Clin Virol. 2010;49:94–9. DOIPubMedGoogle Scholar
- Lau SK, Yip CC, Tsoi HW, Lee RA, So LY, Lau YL, Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, HRV-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–64. DOIPubMedGoogle Scholar