Clinical Significance of Escherichia albertii
Tadasuke Ooka, Kazuko Seto, Kimiko Kawano, Hideki Kobayashi, Yoshiki Etoh, Sachiko Ichihara, Akiko Kaneko, Junko Isobe, Keiji Yamaguchi, Kazumi Horikawa, Tânia A.T. Gomes, Annick Linden, Marjorie Bardiau, Jacques G. Mainil, Lothar Beutin, Yoshitoshi Ogura, and Tetsuya Hayashi

Author affiliations: University of Miyazaki, Miyazaki, Japan (T. Ooka, Y. Ogura, T. Hayashi); Osaka Prefectural Institute of Public Health, Osaka, Japan (K. Seto); Miyazaki Prefectural Institute for Public Health and Environment, Miyazaki (K. Kawano); National Institute of Animal Health, Ibaraki, Japan (H. Kobayashi); Fukuoka Institute of Health and Environmental Sciences, Fukuoka, Japan (Y. Etoh, S. Ichihara, K. Horikawa); Yamagata Prefectural Institute of Public Health, Yamagata, Japan (A. Kaneko); Toyama Institute of Health, Toyama, Japan (J. Isobe); Hokkaido Institute of Public Health, Hokkaido, Japan (K. Yamaguchi); Universidade Federal de São Paulo, São Paulo, Brazil (T.A.T. Gomes); University of Liège, Liège, Belgium (A. Linden, M. Bardiau, J.G. Mainil); Federal Institute for Risk Assessment, Berlin, Germany (L. Beutin)
Main Article
Figure 2
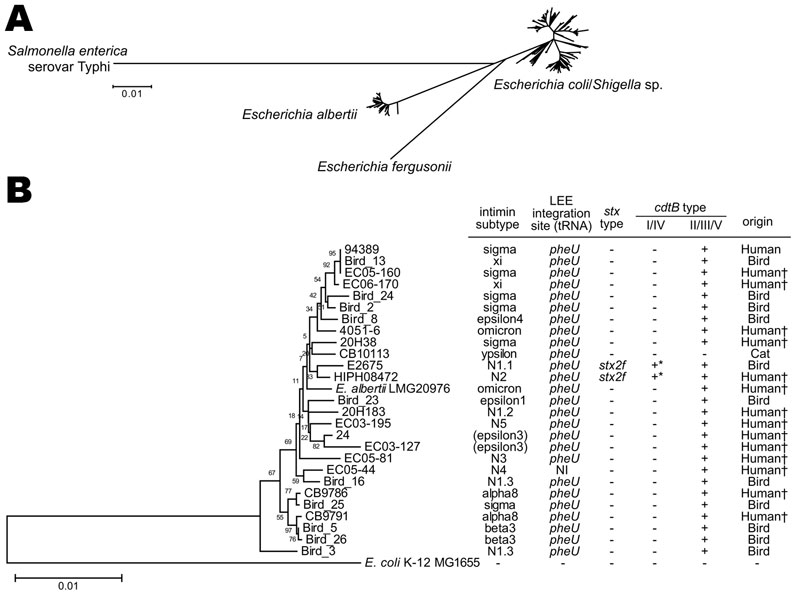
Figure 2. Neighbor-joining tree of 179 eae-positive Escherichia coli and Escherichia albertii strains analyzed by multilocus sequence analysis. The tree was constructed with the concatenated partial nucleotide sequences of 7 housekeeping genes (see Technical Appendix for protocol details). A) The whole image of the 179 strains examined and 10 reference strains (E. coli/Shigella sp., E. fergusonii, and Salmonella enterica serovar Typhi) is shown. B) Enlarged view of the E. albertii lineage and the genetic information of the identified E. albertii strains. E. coli strain MG1655 and E. albertii type strain LMG20976 are included as references. There was no phylogenetic correlation between human and animal isolates. The cdtB genes indicated by * are classified as subtype I. The strains indicated by † were isolated from patients with signs and symptoms of gastrointestinal infection. LEE, locus of enterocyte effacement; NI, not identified; NA, not applicable Scale bars indicate amino acid substitutions (%) per site.
Main Article
Page created: February 15, 2012
Page updated: February 15, 2012
Page reviewed: February 15, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
