Volume 19, Number 6—June 2013
Dispatch
Ciprofloxacin-Resistant Salmonella enterica Serovar Kentucky in Canada
Abstract
We report emergence of ciprofloxacin-resistant Salmonella enterica serovar Kentucky in Canada during 2003–2009. All isolates had similar macrorestriction patterns and were multilocus sequence type ST198, which has been observed in Europe and Africa. Ciprofloxacin-resistant S. enterica serovar Kentucky represents 66% of all ciprofloxacin-resistant nontyphoidal Salmonella sp. isolates observed in Canada since 2003.
Infections with Salmonella spp. are a major health concern for humans and animals on a global scale. Although most cases of salmonellosis result in uncomplicated diarrhea, elderly and immunocompromised persons can be at risk for more severe invasive infections, which can be life-threatening and may require antimicrobial drug therapy (1). The drugs of choice for treating these invasive infections are fluoroquinolones (for adults) or cephalosporins.
One of the main drivers of antimicrobial drug resistance in Salmonella spp. is use of antimicrobial drugs in food-producing animals. For example, high rates of cephalosporin resistance in Salmonella enterica serovar Heidelberg isolated from poultry, retail chicken meat, and humans were observed in Quebec, Canada in 2003. After a voluntary withdrawal of cephalosporins was instituted by the Quebec broiler industry in 2005, rates of ceftiofur resistance dramatically decreased in animals and humans (2).
As with cephalosporin resistance, ciprofloxacin resistance in Salmonella spp. is a growing concern. Recently, S. enterica serovar Kentucky isolates have been described in Europe and Africa that were ciprofloxacin resistant (3). In addition, these isolates were resistant to multiple classes of antimicrobial drugs, which further complicates treatment options for invasive disease. No S. enterica serovar Kentucky isolates submitted to the National Antimicrobial Resistance Monitoring System in the United States were ciprofloxacin resistant (3). The purpose of this study was to describe the epidemiology and characterize isolates of ciprofloxacin-resistant S. enterica serovar Kentucky identified in Canada.
The Canadian Integrated Program for Antimicrobial Resistance Surveillance (CIPARS), established in 2003, monitors antimicrobial drug use and resistance in selected species of enteric bacteria from humans, animals, and animal-derived food sources across Canada (www.phac-aspc.gc.ca/ cipars-picra/surv-eng.php). Human Salmonella isolates were submitted by all provincial public health laboratories in Canada to the National Microbiology Laboratory for further characterization. Antimicrobial drug susceptibility testing was performed by using broth microdilution (Sensititer Automated Microbiology System; Trek Diagnostic Systems Ltd., Westlake, OH, USA) and breakpoints established by the Clinical Laboratory Standards Institute (4).
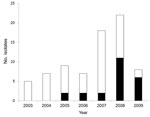
A total of 76 S. enterica serovar Kentucky isolates were submitted to the CIPARS program during 2003–2009, and 23 (30%) isolates showed ciprofloxacin resistance (MIC ≥4 mg/L) during the study (Figure 1). Thirty-five (46%) isolates were susceptible to all antimicrobial drugs tested. Ciprofloxacin-resistant isolates were identified from human case-patients in British Columbia (n = 2), Alberta (n = 2), Saskatchewan (n = 1), Ontario (n = 12), Quebec (n = 5), and Prince Edward Island (n = 1). Age information was available for 54 of 76 case-patients infected with S. enterica serovar Kentucky during the study period.
Of these isolates, 11 (14.5%) were resistant to ciprofloxacin. Ciprofloxacin resistance was observed among case-patients 18–69 years of age, and 5 of 11 were 18–29 years of age. Case-patients 18–29 years of age were 8 times more likely to have a ciprofloxacin-resistant strain than case-patients 50–69 years of age (odds ratio [OR] 8.3, 95% CI 1.034–67.198, p = 0.046). Of 21 ciprofloxacin-resistant isolates from case-patients who reported site of isolation, 20 were identified from feces and 1 from urine. There were no differences in site of isolation between ciprofloxacin-resistant and ciprofloxacin-susceptible S. enterica serovar Kentucky isolates. Although the total number of isolates associated with human infections was rare, of the 21,426 nontyphoidal Salmonella spp. submitted for susceptibility testing as part of the human component of the CIPARS program since 2003, S. enterica serovar Kentucky had a significantly higher rate of ciprofloxacin resistance than all other nontyphoidal Salmonella isolates and comprised 66% (23/35; p<0.0001) of all ciprofloxacin-resistant isolates identified during that period.
In Canada, ciprofloxacin-resistant S. enterica serovar Kentucky was first identified in 2005, when 22% (2/9) of isolates submitted for drug susceptibility testing were resistant to this drug. A significant increase (OR 10.5, 95% CI 1.115–9.913, p = 0.04) in the number of isolates resistant to ciprofloxacin was observed in 2009 compared with results in 2005. The largest number occurred during 2008–2009, when ciprofloxacin-resistant isolates comprised 57% (17/30) of all S. enterica serovar Kentucky isolates identified (Figure 1). The number of cases reported in Canada is comparable with that reported in Denmark over a similar period (3).
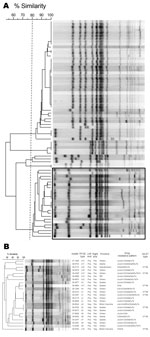
We typed all isolates by using pulsed-field gel electrophoresis as described and restriction enzyme XbaI (5). A dendrogram depicting the results was generated with BioNumerics version 3.5 (Applied Maths, Sint-Martens-Latem, Belgium) and is shown in Figure 2. All ciprofloxacin-resistant isolates clustered with a percentage similarity >80%; only 1 ciprofloxacin-susceptible isolate was found in this cluster (Figure 2, panel A).
Multilocus sequence typing (MLST) was performed on a subset of 8 isolates on the basis of differences in pulsed-field gel electrophoresis patterns and variations in antimicrobial drug resistance. Data were submitted to the MLST database website (http://mlst.ucc.ie/mlst/dbs/Senterica) to determine MLST types (6). All isolates tested were sequence type (ST) 198 (Figure 2, panel B). This sequence type and similar antimicrobial drug resistance patterns have been recently reported in France, England and Wales, Denmark, Belgium, and Africa (3,7,8).
Many ST198 multidrug-resistant isolates observed in Europe and Africa contained Salmonella genomic island 1 (SGI1) variants, particularly, SGI1-K, SGI1-Q, and SGI1-P. To determine whether ciprofloxacin-resistant isolates from Canada harbored similar SGI1 variants, we used PCR to detect the chromosomal left and right junctions of SGI1 as described (9,10). The right junction was found in 22 of 23 isolates, and the left junction was found in 18 of 23 isolates (Figure 2, panel B). Further studies are needed to identify specific SGI variants in the isolates.
Analysis of S. enterica serovar Kentucky isolates obtained during 2003–2009 from animal and retail meat samples as part of CIPARS did not identify any ciprofloxacin-resistant isolates (www.phac-aspc.gc.ca/cipars-picra/index-eng.php). This finding suggests that human infections in Canada were not acquired from domestically produced food. Many ciprofloxacin-resistant S. enterica serovar Kentucky human infections identified in Europe have been linked to travel to countries in Africa (3). Of 23 case-patients in Canada, we obtained travel history for 11. Travel history was defined as previous travel out of Canada within the past 7 days. Four case-patients had traveled to Morocco (1 also had traveled to Spain and Portugal), 3 had traveled to Egypt, 1 had traveled to Libya, and 3 had traveled to Africa (no country reported).
Resistance to ciprofloxacin in Salmonella spp. is a growing concern because it limits the ability to treat invasive disease. In this study, we described the characteristics of ciprofloxacin-resistant S. enterica serovar Kentucky isolates in Canada. Similar drug-resistance patterns and genetic backgrounds of S. enterica serovar Kentucky have been observed in Europe and linked to travel to countries in Africa (3). That most isolates had multidrug resistance phenotypes is of particular concern. Further studies are required to determine risk factors for acquisition of these infections in Canada.
Dr. Mulvey is chief of the Antimicrobial Resistance and Nosocomial Infections Section of the National Microbiology Laboratory at the Public Health Agency of Canada in Winnipeg, Manitoba, Canada. His research interests include the molecular epidemiology of antimicrobial drug–resistant pathogens.
Acknowledgment
This study was supported by the Public Health Agency of Canada.
References
- American Academy of Pediatrics. Salmonella infections. In: Pickering LK, editor. Red book: 2009 report of the committee on infectious diseases, 28th ed. Elk Grove Village (IL): American Academy of Pediatrics; 2009. p. 584.
- Dutil L, Irwin R, Finley R, Ng LK, Avery B, Boerlin P, Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg Infect Dis. 2010;16:48–54. DOIPubMedGoogle Scholar
- Le Hello S, Hendriksen RS, Doublet B, Fisher I, Nielsen EM, Whichard JM, International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J Infect Dis. 2011;204:675–84. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing: eighteenth informational supplement. CLSI document M100–S18. Wayne (PA): The Institute; 2008.
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. DOIPubMedGoogle Scholar
- Harbottle H, White DG, Mcdermott PF, Walker RD, Zhao S. Comparison of multilocus sequence typing, pulsed-field gel electrophoresis, and antimicrobial susceptibility typing for characterization of Salmonella enterica serotype Newport isolates. J Clin Microbiol. 2006;44:2449–57. DOIPubMedGoogle Scholar
- Collard JM, Place S, Denis O, Rodriguez-Villalobos H, Vrints M, Weill F-X, Travel-acquired salmonellosis due to Salmonella Kentucky resistant to ciprofloxacin, ceftriaxone and co-trimoxazole and associated with treatment failure. J Antimicrob Chemother. 2007;60:190–2. DOIPubMedGoogle Scholar
- Weill F-X, Bertrand S, Guesnier F, Baucheron S, Cloeckaert A, Grimont PA. Ciprofloxacin-resistant Salmonella Kentucky in travelers. Emerg Infect Dis. 2006;12:1611–2. DOIPubMedGoogle Scholar
- Boyd D, Peters GA, Cloeckaert A, Boumedine KS, Chaslus-Dancla E, Imberechts H, Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J Bacteriol. 2001;183:5725–32. DOIPubMedGoogle Scholar
- Doublet B, Praud K, Bertrand S, Collard J-M, Weill F-X, Cloeckaert A. Novel insertion sequence- and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob Agents Chemother. 2008;52:3745–54. DOIPubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 19, Number 6—June 2013
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Michael R. Mulvey, National Microbiology Laboratory, 1015 Arlington St, Winnipeg, Manitoba, R3E 3R2, Canada
Top