Volume 20, Number 1—January 2014
Research
Human Parechovirus Infection, Denmark
Figure 2
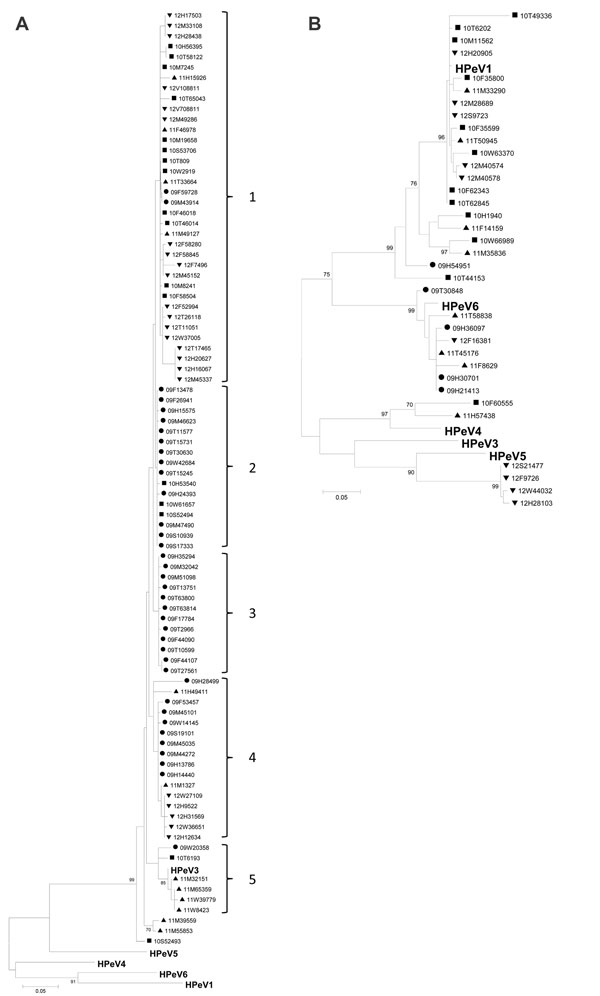
Figure 2. . Phylogenetic analysis of the viral protein (VP) 3 (A) and VP1 (B) nucleotide sequences of human parechoviruses (HPeV), Denmark, January 2009–December 2012. Maximum-likelihood analysis of HPeVs detected in 2009 are indicated by dots, 2010 by squares, 2011 by triangles, and 2012 by inverted triangles. Reference sequences from GenBank representing the different HPeV genotypes identified in this study are shown in boldface. The following sequences were used as references: HPeV1, GenBank accession no. JX575746; HPeV3, JX826607; HPeV4, AB433629; HPeV5, JX050181; HPeV6, AB25282. Scale bars indicate nucleotide substitutions per site.
Page created: January 03, 2014
Page updated: January 03, 2014
Page reviewed: January 03, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.