Volume 20, Number 8—August 2014
Dispatch
Isolation of MERS Coronavirus from a Dromedary Camel, Qatar, 2014
Abstract
We obtained the full genome of Middle East respiratory syndrome coronavirus (MERS-CoV) from a camel in Qatar. This virus is highly similar to the human England/Qatar 1 virus isolated in 2012. The MERS-CoV from the camel efficiently replicated in human cells, providing further evidence for the zoonotic potential of MERS-CoV from camels.
Middle East respiratory syndrome coronavirus (MERS-CoV) is a novel coronavirus that can cause severe lower respiratory tract infection in humans (1,2). MERS-CoV clusters with viruses in the genus Betacoronavirus; the closest relative to this virus is bat CoVs clade 2c (3). Although bats are believed to carry different CoV ancestors, antibody reactivity against MERS-CoV has been found in serum from dromedary camels from countries within the Arabian Peninsula (4–7), Egypt (8), and the Canary Islands (4). More recently, MERS-CoVs that phylogenetically cluster with human MERS-CoVs were detected in camels from Qatar, Saudi Arabia, and Egypt (7,9–12). To further characterize MERS-CoV from camels, we screened nose swab samples from camels in Qatar.
In February 2014, nasal swab samples were collected from 53 healthy dromedary camels in Doha, Qatar. After sampling, swabs were put into tubes containing viral transport medium and stored at −80°C until shipment to the Netherlands on dry ice, as described (9).
Total nucleic acids from nasal swabs were isolated by using the MagnaPure 96 total nucleic acid isolation kit (Roche, Mannheim, Germany), and samples were tested for MERS-CoV by using 2 TaqMan assays: 1 for the envelope (upE) and 1 for the nucleocapsid gene (N), as described previously (9,13). In each assay we detected MERS-CoV RNA in a sample from an 8-month-old camel. The cycle threshold of the positive sample was 12.9 in the upE assay and 11.3 in the N assay.
For further genomic characterization, RNA was isolated from 50 μL of 1 swab sample with the QIAamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany), eluted in 60 μL water, and reverse transcribed with the Superscript III First-Strand Synthesis System (Life Technologies (Bleiswijk, the Netherlands) with random hexamers. The MERS-CoV genome was amplified by using MERS-CoV–specific overlapping primer sets as described previously (3). Amplified MERS-CoV fragments were sequenced directly on both strands by using the BigDye Terminator version 3.1 Cycle Sequencing kit on an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Bleiswijk, the Netherlands). To obtain the 5′ and 3′ ends, we used the FirstChoice RLM-RACE kit (Ambion, Bleiswijk, the Netherlands) according to the manufacturer’s protocols. Using overlapping sequence fragments, we assembled the complete MERS-CoV genome, except for 1 nt potentially missing at the 5′ end.
The genome was 30,117 nt long, including 12 nt at the 3′ poly A tail (MERS-CoV camel/Qatar_2_2014, GenBank accession no. KJ650098). Similar to the genome of human MERS-CoV isolates, the genome of camel MERS-CoV isolates contains 10 complete open reading frames (ORFs) (ORF 1ab, spike, ORF3, ORF4a, ORF4b, ORF5, envelope, membrane, nucleocapsid, and ORF8b), 8 transcription-regulatory sequences, and 2 terminal untranslated regions. The alignment of the camel MERS-CoV with known human MERS-CoVs, including 1 near-complete camel MERS-CoV (NRCE_HKU205) sequence, showed overall nucleotide identities of 99.5%–99.9% between camel and human MERS-CoV isolates from different geographic regions.
Phylogenetic analysis of the complete genome clearly showed that MERS-CoV camel/Qatar_2_2014 is highly similar to human MERS-CoV; the closest relative to camel MERS-CoV was England/Qatar1 2012 (99.9% identity) (Figure 1), and it was clearly distinct from the camel MERS-CoV (99.5% identity) isolated from camels at a different location in Qatar and in Egypt (9,11). Comparison of spike protein amino acid sequences from various human and camel isolates showed that this protein is highly conserved between this camel virus and other human isolates (online Technical Appendix Table, wwwnc.cdc.gov/EID/article/20/8/14-0663-Techapp1.pdf).
In addition, most amino acid residues critical for receptor binding (14) are identical in human and camel isolates, except for L506F in England/Qatar1. The biologic relevance of this mutation has not been investigated. The presence of arginine at position 1020 in the camel virus isolate might indicate that selective pressure at this site has probably not taken place as previously postulated. The fact that a MERS-CoV from a camel is highly similar to that from a human patient who probably became infected >1 year earlier in the same region suggests that this virus is maintained within camel populations and further supports the hypothesis that MERS-CoV can be transmitted from camels to humans.
To test for the presence of infectious virus, we titrated the swab sample on Vero cells (ATCC no. CCL-81). After 48 hours, we observed cytopathic changes in cells (320 50% tissue culture infectious dose/mL). After isolation, the passage-3 virus stock was used for all subsequent experiments.
To check for adaptive mutations obtained during cell culture, we used 454 deep-sequencing technology (Roche, Indianapolis, IN, USA) to analyze the full-genome sequence as described elsewhere (3). A total of 57,655 sequence reads were obtained, of which 17,056 were specific for MERS-CoV, revealing ≈99.77% of the virus genome. Genome coverage ranged from 1 to 2,082 reads at single nucleotide positions. Gaps or regions with coverage of <4 reads were confirmed by Sanger sequencing. When the genome of the passaged virus was aligned with the genome of the initial clinical isolate, we did not observe any mutations acquired during passaging.
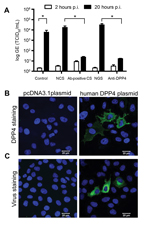
To further functionally characterize this virus isolate, we subsequently inoculated human hepatoma (Huh-7) cells with MERS-CoV camel/Qatar_2_2014. After 2 days, virus-induced cytopathic effects were observed in the inoculated cell cultures (Technical Appendix Figure). In addition, a strong increase in virus titer was measured in the cell supernatant (Figure 2, panel A); produced virus could be passaged (not shown). Virus production in Huh-7 cells was blocked by preincubating camel MERS-CoV with a 1:200 dilution of serum from MERS-CoV antibody–positive camels (9) but not with seronegative camel serum (4) (Figure 2, panel A). Infection of Huh-7 cells could also be blocked by preincubation of cells with polyclonal antiserum against human DPP4 but not with control serum (Figure 2, panel A). Furthermore, transfection of nonsusceptible MDCK cells with human DPP4 (Figure 2, panel B), but not with empty vector, conferred susceptibility to infection with camel MERS-CoV (Figure 2, panel C). These data demonstrate that the MERS-CoV obtained from a dromedary camel is able to replicate in human cells and uses DPP4 as entry receptor, similar to MERS-CoV isolates obtained from human patients (15).
We isolated MERS-CoV from the nasal cavity of 1 dromedary camel and demonstrated its infectiousness. Further studies are needed to test whether camels infected at a young age are more likely than adult dromedary camels to excrete infectious virus, possibly because of the MERS-CoV seronegative status of the younger camels. In addition, our results add to recent findings that MERS-CoVs from camels and humans are nearly identical (9–11). As might be expected from the high level of conservation in the critical interacting amino acids in the receptor-binding domain of the camel and human MERS-CoV isolates (online Technical Appendix Table), we show that camel MERS-CoV can infect human Huh-7 cells by using the same entry receptor as the human MERS-CoV isolates (15). Collectively, combined with the observation that the sequence of this virus was most closely related to that of a virus from a human patient who acquired MERS-CoV in Qatar a year earlier, these data support the hypothesis that dromedary camels are a reservoir for MERS-CoV and can transmit the infection to humans. However, whether exposure of humans to camels directly can lead to human infection cannot be concluded from our data. We are not aware of a connection between the camel population sampled in this study and the patient infected with MERS-CoV England/Qatar 1. Future epidemiologic studies are needed to investigate whether contact with camels or camel products constitutes a risk factor for MERS-CoV infection.
Dr Stalin Raj is a postdoctoral scientist at the Department of Viroscience, Erasmus Medical Center, Rotterdam, the Netherlands. His research interests are the molecular characterization and pathogenesis of emerging viruses.
Acknowledgments
We are grateful to the Joint Supreme Council of Health and Animal Resources Department of Ministry of Environment field investigation team for exceptional research assistance; to the Doha Camel slaughterhouse veterinarians, staff, and workers for their help; and to the Supreme Council of Health Administration for funding this study through a grant from the Health Promotion Department and the Communicable Disease Control Department routine budget. We also thank Thomas P.Samuel, Redentor Cuizon, Ronald R. Manaor, Khalid Yousif, and Farid Abdoudia for help with collecting samples.
This work was funded by a grant from the Dutch Scientific Research (no. 40-00812-98-13066), the European Community's Seventh Framework Program (FP7/2007–2013) under the project “European Management Platform for Emerging and Re-emerging Infectious disease Entities”; European Commission (agreement no. 223498); and the project “Anticipating Global Onset of Epidemics” (agreement no. 102938).
References
- Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–20. DOIPubMedGoogle Scholar
- World Health Organization, MERS-CoV Research Group. State of knowledge and data gaps of Middle East respiratory syndrome coronavirus (MERS-CoV) in humans. PLoS Curr. 2013;5:pii: ecurrents.outbreaks.0bf719e352e7478f8ad85fa30127ddb8
- van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBiol. 2012;3:e00473–12.PubMedGoogle Scholar
- Reusken CB, Haagmans BL, Muller MA, Gutierrez C, Godeke GJ, Meyer B, Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–66. DOIPubMedGoogle Scholar
- Hemida MG, Perera R, Wang P, Alhammadi MA, Siu LY, Poon LL, Middle East respiratory syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveill. 2013;18:20659 .PubMedGoogle Scholar
- Meyer B, Müller MA, Corman VM, Reusken CBEM, Ritz D, Godeke G-D. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis. 2014;20:552–9. DOIPubMedGoogle Scholar
- Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, de Wit E, Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. MBio. 2014;5:e00884–14 .DOIPubMedGoogle Scholar
- Perera RA, Wang P, Gomaa MR, El-Shesheny R, Kandeil A, Bagoto O, Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18:20574 .PubMedGoogle Scholar
- Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–5. Epub 2013 Dec 17. DOIPubMedGoogle Scholar
- Memish ZA, Cotton M, Meyer B, Watson SJ, Alsahafi AJ, Al Rabeeah AA, Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis. 2014. [Epub ahead of print].
- Chu DKW, Poon LLM, Gomaa MM, Shehata MM, Perera RAPM, Zeid DAE. MERS coronaviruses in dromedary camels, Egypt. Emerg Infect Dis. 2014 [Epub ahead of print]. DOIGoogle Scholar
- Hemida MG, Chu DKW, Poon LLM, Perera RAPM, Alhammadi MA, Ng H-Y, MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis. 2014 [Epub ahead of print].
- Corman VM, Eckerle I, Bleicker T, Zaki A, Landt O, Eschbach-Bludau M, Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285 .PubMedGoogle Scholar
- Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–31. Epub 2013 Jul 7. DOIPubMedGoogle Scholar
- Raj VS, Mou H, Smits SL, Dekkers DH, Müller MA, Dijkman R, Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–4. DOIPubMedGoogle Scholar
Figures
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 20, Number 8—August 2014
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Bart L. Haagmans, Department of Viroscience, Erasmus Medical Center, P.O. Box 2040, 3000 CA Rotterdam, the Netherlands; ; Mohd M. AlHajri, Supreme Council of Health, PO Box 42, Doha, Qatar
Top