Volume 21, Number 9—September 2015
THEME ISSUE
Emerging Infections Program
Emerging Infections Program
Foodborne Diseases Active Surveillance Network—2 Decades of Achievements, 1996–2015
Abstract
The Foodborne Diseases Active Surveillance Network (FoodNet) provides a foundation for food safety policy and illness prevention in the United States. FoodNet conducts active, population-based surveillance at 10 US sites for laboratory-confirmed infections of 9 bacterial and parasitic pathogens transmitted commonly through food and for hemolytic uremic syndrome. Through FoodNet, state and federal scientists collaborate to monitor trends in enteric illnesses, identify their sources, and implement special studies. FoodNet’s major contributions include establishment of reliable, active population-based surveillance of enteric diseases; development and implementation of epidemiologic studies to determine risk and protective factors for sporadic enteric infections; population and laboratory surveys that describe the features of gastrointestinal illnesses, medical care–seeking behavior, frequency of eating various foods, and laboratory practices; and development of a surveillance and research platform that can be adapted to address emerging issues. The importance of FoodNet’s ongoing contributions probably will grow as clinical, laboratory, and informatics technologies continue changing rapidly.
During the late 1980s and early 1990s, recognizing inconsistencies inherent in passive national surveillance systems, epidemiologists at the Centers for Disease Control and Prevention (CDC) proposed creating a population-based active surveillance system to better measure the frequency of enteric infections and their effects on society. However, resources for these improvements were not available. Then, in late 1992 and early 1993, hamburger patties contaminated with Escherichia coli O157 caused 732 laboratory-confirmed infections and the deaths of 4 children. After this outbreak, the US Department of Agriculture (USDA) implemented a risk-based meat inspection system. Public health and regulatory officials needed a method to determine whether the changes made by regulatory agencies and the industry were followed by declines in infections. The outbreak had focused attention on the need for reliable data on the incidence of infections caused by enteric pathogens; changes in the incidence over time; and estimates of the actual numbers of illnesses, hospitalizations, and deaths they cause. Therefore, in 1995, with support from Food Safety and Inspection Service (FSIS) of the USDA, CDC established the Foodborne Diseases Active Surveillance Network (FoodNet), an active, population-based sentinel surveillance system. FoodNet monitors changes in the incidence of selected major bacterial and parasitic illnesses transmitted commonly by food, attributes illnesses to sources and settings, and estimates the total numbers of foodborne illnesses in the United States.
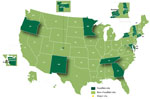
FoodNet, a core part of CDC’s Emerging Infections Program, is a collaboration among CDC, 10 state health departments, USDA-FSIS, and the Food and Drug Administration (FDA). Over time, the surveillance area has grown to include ≈48 million persons (≈15% of the US population) in Connecticut, Georgia, Maryland, Minnesota, New Mexico, Oregon, and Tennessee and in selected counties in California, Colorado, and New York (1) (Figure 1). More information about the program and its activities is available at http://www.cdc.gov/FoodNet. The cost of funding for the surveillance sites and CDC, typically <$7 million per year, is dwarfed by the economic impact of the illnesses monitored. Salmonella infections alone cost ≈$3.6 billion each year in direct medical costs, productivity, and years of potential life lost (2).
The community of multidisciplinary FoodNet scientists collaborates to track infections transmitted commonly by food and to study the sources of infections. FoodNet’s annual report of confirmed infections caused by major pathogens, published within months of the end of each calendar year, is sometimes referred to as the foodborne illness “report card” for the nation. Public health officials, regulatory agencies, and industry use it to gauge progress in food safety and to determine when new policies and prevention efforts are needed (3).
FoodNet’s major contributions include the establishment of reliable, active population-based surveillance of enteric diseases; development and implementation of epidemiologic studies to determine risk and protective factors for sporadic enteric infections; population and laboratory surveys that describe the features of gastrointestinal illnesses, medical care–seeking behavior, food eating patterns, and laboratory practices; and development of a surveillance and research platform that can be adapted to address emerging issues (Table). It is the only US system focused on obtaining comprehensive information about sporadic infections caused by pathogens transmitted commonly through food.
Active Surveillance
FoodNet’s core activity is active surveillance for laboratory-confirmed bacterial infection caused by Campylobacter, Listeria, Salmonella, Shiga toxin–producing Escherichia coli (STEC) O157 and non-O157, Shigella, Vibrio, and Yersinia and parasitic infection caused by Cryptosporidium and Cyclospora. FoodNet does not track agents for which clinical laboratories do not routinely test (e.g., norovirus). As laboratory practices change, the surveillance system adapts. During the 2000s, culture-independent diagnostic tests (CIDTs) became commercially available to detect Shiga toxin. Recognizing that some laboratories might stop culturing for STEC O157, leading to a perceived decline in infections, FoodNet began surveillance for hemolytic uremic syndrome (HUS) as another way to track STEC O157 infections. Most cases of HUS are caused by STEC O157. As more clinical laboratories began using CIDTs to detect other pathogens, FoodNet responded by gathering data on laboratory practices and on pathogens detected by these tests and by encouraging reflex culturing of specimens that test positive (4).
FoodNet staff at each site receive reports of every identification of a pathogen under surveillance from clinical laboratories that conduct tests on patients’ specimens ordered by health care providers. They conduct periodic audits to ensure that all pathogens identified are reported (3). In 1999, to ensure the validity of data summarized across all sites, FoodNet developed and began tracking metrics related to reporting, a process unusual for CDC programs at that time. For each person with an infection, FoodNet staff collect demographic information and determine whether the person was hospitalized and whether he or she survived. In 2004, FoodNet began collecting data on whether the infection was part of an outbreak and whether the patient had traveled internationally.
FoodNet has the flexibility to conduct special surveillance projects in response to concerns about emerging infectious diseases, as when variant Creutzfeldt-Jakob disease emerged (3). In 2010, FoodNet conducted surveillance for Cronobacter sakazakii infections and found infection in all age groups, the highest rate of invasive infections in infants, and data suggesting that urine might be a more common site of infection than previously thought (5).
Tracking Incidence and Changes Over Time
FoodNet tracks the incidence of infections and HUS to assess the effectiveness of measures aimed at preventing infections and to monitor progress toward national health goals. To measure changes over time and minimize the spurious effect of annual fluctuations, FoodNet has used 2 baseline periods of 3 consecutive years each. The first, 1996–1998, is the initial 3 years of surveillance; the second, 2006–2008, was used to develop the US Department of Health and Human Services Healthy People 2020 goals (6). In 2008, FoodNet began also reporting changes from the average annual incidence for the 3 years preceding the year of the report. In 2012, FoodNet began reporting a measure of overall change in the incidence of bacterial foodborne illness. This measure combines data for infections caused by the 6 bacterial pathogens monitored by the network for which >50% of illnesses are estimated to be transmitted by food (7). To account for variations in the surveillance area, FoodNet uses a main-effects log-linear Poisson (negative binomial) regression model to assess changes in incidence rates (1).
Each spring, FoodNet summarizes preliminary data and changes in incidence for the preceding year (Figure 2) in CDC’s Morbidity and Mortality Weekly Report. Public health officials, regulatory agencies, industry, and consumer groups use these data to assess the effect of food safety interventions (3). FoodNet has documented significant decreases in the incidence of E. coli O157 infections since 1996–1998 and in HUS since 2001, supporting other data indicating that regulatory and industry actions have made ground beef safer (8). FoodNet also has documented lack of significant change in the overall incidence of Salmonella infections and marked changes in some specific serotypes, indicating that efforts targeting specific serotypes are needed to decrease Salmonella infections. In response to these findings and to recent outbreaks, FSIS created performance standards mandating the upper limit of allowable Salmonella contamination of chicken parts (8). Poultry is also a major source of Campylobacter infections (9). In 2011, in response to FoodNet data showing little progress in reducing these infections, FSIS issued the first performance standards that limited the allowable contamination of chicken and turkey with Campylobacter (10).
FoodNet data are used to guide the development of and monitor progress toward national goals and health objectives, such as the US Department of Health and Human Services’ high priority goal to reduce S. enterica serotype Enteritidis infections from eggs after implementation of the Egg Safety Rule that was passed in 2009 (6). FoodNet data also are used to monitor progress on 7 illnesses included in the Healthy People National Health Objectives.
Determining Sources and Outcomes of Infections
Understanding sources and settings of illnesses informs the development of recommendations, regulations, and interventions to reduce illnesses. FoodNet collects data to determine the relative importance of various routes of infection, including nonfood sources. Kendall et al. reported that, with wide variation by pathogen, 13% of persons infected with a pathogen monitored by FoodNet had recently traveled internationally; the authors identified regions to which travel carries the highest risk for illness (11). Hale et al. used data from FoodNet and other sources to estimate that 13% of illnesses caused by 7 enteric pathogens were attributable to contact with animals and their environments (12).
FoodNet has identified numerous risky food, environment, and animal exposures through case–control studies of sporadic Campylobacter, Cryptosporidium, Listeria, Salmonella, and STEC O157 infections, described in 19 journal articles (more information available at http://www.cdc.gov/FoodNet). It is currently conducting a case–control study of non-O157 STEC infections to assess risk factors and correlate virulence factors with symptoms. These studies have yielded rich data of long-term value. When the FDA needed data on sources of S. enterica serotype Enteritidis illnesses before the Egg Rule was implemented, Gu et al. reanalyzed data from an old FoodNet case–control study with a new method and determined that egg-related exposures had the highest attributable fraction (13). A case–control study of Listeria infections showed an unexpected association with eating melons (14). In response, CDC modified the Listeria Initiative questionnaire used to interview patients with Listeria infection. As a result, when Colorado detected a large Listeria outbreak, investigators already had information about cantaloupe consumption from many patients, and the melons were more quickly implicated and removed from the market (15). Case–control studies are resource-intensive and so are conducted infrequently. In 2014 FoodNet began routinely collecting exposure data from patients with some Salmonella infections and is exploring ways to use these data in models that attribute illnesses to sources.
Population and Laboratory Surveys
FoodNet conducted 5 population surveys beginning in 1996, with only a 2-year gap before the last survey ended in 2007. In addition to obtaining data for estimating illnesses (described in the next section), the surveys asked participants how recently they ate selected foods. These data have been used for many analyses. Shiferaw et al. found a higher proportion of men reported eating pink hamburger and runny eggs, whereas a higher proportion of women ate fruits and vegetables (16). The population surveys have been used frequently in outbreak investigations. Epidemiologists compare frequencies of specific exposures reported by outbreak patients with those of a comparable population in the survey to quickly generate, confirm, or refute hypotheses about sources of illness. The ready availability of these data saves time over traditional methods of finding controls (e.g., by random-digit telephone calls, followed by interviews of persons reached who agree to participate) (17). During a 2012–2013 multistate outbreak of S. enterica serotype Heidelberg infections, 79% of patients reported eating chicken in the week before illness began, significantly higher than the 65% reported in the 2006–2007 FoodNet population survey (18). That finding, with other epidemiologic, laboratory, and traceback findings, helped link the outbreak to chicken from 1 producer. The population surveys also have provided a platform for obtaining information quickly in a crisis. When bovine spongiform encephalopathy emerged as a public health concern during the mid-2000s, questions about hunting practices, eating venison, and travel to countries in which bovine spongiform encephalopathy had been reported in animals were added to the 2006–2007 survey (19).
FoodNet conducts surveys of clinical laboratories to determine practices. By analyzing data from surveys conducted in 1995, 1997, and 2000, Voetsch et al. determined that variations in laboratory practice by site might explain some of the observed differences in the incidence of STEC O157 infection (20). A survey in 2005 found that adherence to recommendations for isolation and identification of Campylobacter varied substantially among laboratories (21). A survey in 2007 showed that most laboratories complied with recommendations for testing STEC O157 but not with recommendations for non-O157 STEC (22). The laboratory surveys provided essential information for estimating the true number of enteric infections (23). In 2012, because of rapidly changing clinical laboratory practices, FoodNet began conducting a survey annually. The 2014 survey showed that CIDT methods were used most often to detect Campylobacter and STEC (4).
Estimating Actual Foodborne and Acute Gastrointestinal Illnesses, Hospitalizations, and Deaths
FoodNet data are central to estimating the numbers of US foodborne illnesses, hospitalizations, and deaths (23,24). Regulatory agencies and lawmakers use these estimates to help decide how to allocate resources for prevention. Mead et al. published the first estimates in 1999 in Emerging Infectious Diseases (24) using early active surveillance data from FoodNet and data from other sources. By 2010, this article was the most frequently cited article published in this journal. After these estimates were published, FoodNet began addressing data gaps and developing improved methods, resulting in revised comprehensive estimates published by Scallan et al. in 2011 (23). Major improvements resulted from the availability of data from >5 times more respondents to population surveys and more detailed information about illnesses reported in those surveys. For both the 1999 and the 2011 estimates, these surveys provided key data on the severity of illnesses, medical care–seeking behavior, and specimen submission. These data and data from FoodNet surveys of laboratories were used to estimate the total number of illnesses for every reported laboratory-confirmed illness of each pathogen. The surveys also provided essential information about the rate of acute gastroenteritis illnesses, which was used to estimate illnesses caused by viral pathogens and by unknown agents (25). An important advancement in the 2011 article was separate estimation of the numbers of illnesses acquired domestically and during international travel, enabled by FoodNet’s collection of information about recent international travel. These new foodborne illness estimates formed the basis for a ground-breaking analysis estimating the number of illnesses attributed to specific food categories (26).
Other Contributions
Linking FoodNet to the National Antimicrobial Resistance Monitoring System (NARMS) has expanded the impact of both surveillance systems. A FoodNet case–control study linked fluoroquinolone-resistant Campylobacter infections with eating poultry at a commercial establishment and with international travel (27). Data from this study contributed to the body of evidence that led FDA to withdraw approval for the use of fluoroquinolones in poultry (28). Krueger et al. conducted a joint FoodNet–NARMS study that showed bloodstream infection was more common among patients infected with resistant than susceptible Salmonella strains (29). By linking FoodNet and NARMS data, Shiferaw et al. found that Shigella isolates from Hispanics and recent international travelers were more likely than other isolates to be resistant to trimethoprim/sulfamethoxazole (30). In another study linking these 2 databases, O’Donnell et al. found that two thirds of persons with S. enterica serotype Enteritidis infections resistant to nalidixic acid had recently traveled internationally (31).
The ability to geocode FoodNet surveillance data and link it to census data has increased FoodNet’s ability to examine health disparities. By geocoding Campylobacter cases and linking to census tract socioeconomic status (SES) measures, Bemis et al. found the incidence of campylobacteriosis in Connecticut increased as neighborhood SES increased except among children <10 years old, for whom incidence increased as SES decreased (32).
The benefits of FoodNet for public health are far-reaching. As experts in surveillance, FoodNet site epidemiologists are often leaders in conducting multistate outbreak investigations, many of which result in industry or regulatory changes that make food safer. Examples include an outbreak of Salmonella infections linked to pot pies (33), an outbreak of E. coli O157 infections linked to spinach (34), and the outbreak of Listeria infections linked to cantaloupe (15). FoodNet site personnel also train local public health nurses, epidemiologists, sanitarians, and laboratorians about foodborne disease surveillance, outbreak detection, investigation, and response. Training helps local public health professionals recognize outbreaks and maintain skills and knowledge needed to respond appropriately.
FoodNet’s influence reaches beyond the United States. Australia’s OZFoodNet and FoodNet-Canada have modeled some of their activities on FoodNet surveillance, and FoodNet staff have collaborated with scientists from other countries to compare the prevalence of diarrheal illness (35). FoodNet scientists have been active in the World Health Organization Global Foodborne Infections Network, which works to enhance the capacity of countries to detect, respond to, and prevent foodborne and other enteric infections.
The importance of FoodNet’s ongoing contributions toward developing epidemiologic methods for assessing diseases transmitted commonly by food likely will grow as clinical, laboratory, and informatics technologies continue changing at a rapid pace. Recent and ongoing advances in CIDTs and molecular diagnostics affect FoodNet surveillance. FoodNet’s responsiveness to this changing landscape is informing ongoing modifications of national surveillance definitions that CDC and all US states use.
For the near future, although CIDTs serve clinical needs, bacterial isolates remain essential for the molecular subtyping and antimicrobial drug susceptibility testing needed for epidemiologic monitoring, outbreak detection, and public health investigations. Public health laboratories in FoodNet states could become key sites for maintaining public health access to isolates of enteric pathogens obtained by reflex culturing after a positive CIDT. Maintaining access to traditional laboratory methods also is necessary to validate and interpret new technologies. Traditional laboratory methods might be needed to help evaluate the significance of detection using highly sensitive genetic techniques of multiple pathogens in a single specimen. Because FoodNet surveillance is built on clinical and public health laboratory diagnosis, laboratories must have the resources required to meet surveillance needs.
As laboratories adopt whole-genome sequencing to identify and characterize enteric pathogens, the ability to identify subtypes associated with particular reservoirs and particular food sources will increase. Detailed epidemiologic data on exposures of ill persons will be needed to make these associations. At this time, although many state and local health departments obtain exposure information, FoodNet surveillance captures only a limited amount. Obtaining more will involve duplicate data entry or designing information technology systems that can interface with a variety of databases housed at local and state health departments and at CDC.
The advent of CIDTs offers opportunities to conduct surveillance for enteric pathogens not monitored now. Some CIDTs can detect enterotoxigenic E. coli infection, which is an important cause of diarrhea in returning travelers and has caused domestic outbreaks (36). The large proportion of the US food supply that is imported, including many fruits and vegetables that are eaten raw, provides opportunities for exposure to pathogens from all over the world. Expanding surveillance to enterotoxigenic E. coli and other pathogens, after clinical laboratories begin detecting them, could lead to greater insight into the causes and sources of enteric infections in the United States and abroad.
FoodNet population surveys have proven valuable as sources of data about rates and severity of acute gastrointestinal illnesses and medical care–seeking and about food eaten and other exposures among well persons in the community (16,37,38). The lack of a population survey after 2007 means that data needed to update estimates of the impact of illness are not available. Although the frequency that various foods are eaten may have changed, these data are still used because up-to-date data are not readily available from other sources. FoodNet is working with its partners to find ways to fund and conduct more frequent surveys.
Awareness is increasing of the need to combat antimicrobial drug resistance. Four of the 18 threats that CDC reported in Antibiotic Resistance Threats in the United States, 2013, are tracked in FoodNet (39).The National Strategy for Combating Antibiotic Resistant Bacteria, announced in 2014 (40), aims to slow the emergence of resistant bacteria and prevent the spread of resistant infections. Surveillance data are needed to determine whether strategies to preserve the effectiveness of antimicrobial drugs are working and whether new threats are emerging. FoodNet’s longstanding collaboration with NARMS likely will increase further to meet this need.
CDC’s information technology method for obtaining surveillance data from FoodNet sites needs updating, including developing the ability to obtain and analyze data by person and to interface with national surveillance systems. FoodNet creates a separate record for each illness diagnosed by the detection of a pathogen; ways to link the record to the ill person are needed to determine whether a person has a co-infection or has sequential illnesses with several pathogens. The existence of a variety of methods for reporting infections to CDC is an ongoing challenge for state health officials. The use of different identifiers for information about the same isolate or illness reported to various CDC surveillance systems (e.g., FoodNet, PulseNet, NARMS) is an obstacle to fully understanding the features of reported infections. FoodNet staff will be engaged in efforts to ensure that national surveillance systems are designed to meet the needs of both states and CDC and that they enable accurate and timely analysis and release of FoodNet data.
The widespread growth of electronic health records in the clinical community presents challenges and opportunities for public health. CDC’s Emerging Infections Program is developing informatics capacity to incorporate data streams from electronic health records, electronic laboratory reporting, and other sources of “big data” (e.g., administrative claims data, social media). FoodNet databases at the sites and CDC are increasingly conforming to national data standards, which will facilitate linking to meaningful use of certified electronic health records technology. Rapid access to clinical data will improve surveillance and epidemiologic studies. Informatics capacity is essential for linking FoodNet surveillance data with geographic information systems and other public health databases (e.g., hospital discharge data, vital statistics, NARMS surveillance data). Lessons learned from pilot projects conducted at FoodNet sites could provide an important foundation for developing public health informatics infrastructure nationally. Through multiagency partnership and collaboration, FoodNet has helped improve food safety in the United States in multiple ways. The surveillance network, which began as a project, provides key data for public health analyses and decision-making, and has become an integral part of CDC’s work. FoodNet has matured and transformed over the last 20 years and continues to evolve. Changes in diagnostic practices that affect surveillance and the need for more detailed and precise information about the major sources of infections and how they change over time are just a few of the issues FoodNet will address during the next decade.
Dr. Henao leads FoodNet in the Enteric Diseases Epidemiology Branch of the Division of Foodborne, Waterborne, and Environmental Diseases, CDC. Her research interests include enteric diseases; epidemiologic research methods; and collection, analysis and interpretation of surveillance data.
Acknowledgment
We thank Alicia Cronquist, Mary Patrick, Paul Cieslak, and Kirk Smith for assistance in developing the concept for the manuscript; Barbara Mahon, Elaine Scallan, Robert Tauxe, and Sue Swensen for helpful suggestions on the manuscript; and members of FoodNet of the Emerging Infections Program for their contributions to the network.
References
- Henao OL, Scallan E, Mahon B, Hoekstra RM. Methods for monitoring trends in the incidence of foodborne diseases: Foodborne Diseases Active Surveillance Network 1996–2008. Foodborne Pathog Dis. 2010;7:1421–6. DOIPubMedGoogle Scholar
- Hoffman S, Maculloch B, Batz M. Economic burden of major foodborne illnesses acquired in the United States. Economic Information Bulletin no. 140. May 2015 [cited 2015 Jun 9]. http://www.ers.usda.gov/media/1837791/eib140.pdf
- Scallan E. Activities, achievements, and lessons learned during the first 10 years of the Foodborne Diseases Active Surveillance Network: 1996–2005. Clin Infect Dis. 2007;44:718–25. DOIPubMedGoogle Scholar
- Iwamoto M, Huang JY, Cronquist AB, Medus C, Hurd S, Zansky S, Bacterial enteric infections detected by culture-independent diagnostic tests—FoodNet, United States, 2012–2014. MMWR Morb Mortal Wkly Rep. 2015;64:252–7 .PubMedGoogle Scholar
- Patrick ME, Mahon BE, Greene SA, Rounds J, Cronquist A, Wymore K, Incidence of Cronobacter spp. infections, United States, 2003–2009. Emerg Infect Dis. 2014;20:1520–3 . DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 1996–2012. MMWR Morb Mortal Wkly Rep. 2013;62:283–7 .PubMedGoogle Scholar
- Henao OL, Crim SM, Hoekstra RM. Calculating a measure of overall change in the incidence of selected laboratory-confirmed infections with pathogens transmitted commonly through food in the Foodborne Diseases Active Surveillance Network (FoodNet), 1996–2010. Clin Infect Dis. 2012;54(Suppl 5):S418–20. DOIPubMedGoogle Scholar
- Crim SM, Griffin PM, Tauxe R, Marder EP, Gilliss D, Cronquist AB, Preliminary incidence and trends of infection with pathogens transmitted commonly through food—Foodborne Diseases Active Surveillance Network, 10 US sites, 2006–2014. MMWR Morb Mortal Wkly Rep. 2015;64:495–9 .PubMedGoogle Scholar
- Friedman CR, Hoekstra RM, Samuel M, Marcus R, Bender J, Shiferaw B, Risk factors for sporadic Campylobacter infection in the United States: a case–control study in FoodNet sites. Clin Infect Dis. 2004;38(Suppl 3):S285–96. DOIPubMedGoogle Scholar
- US Department of Agriculture-Food Safety Inspection Service. New performance standards for Salmonella and Campylobacter in young chicken and turkey slaughter establishments: response to comments and announcement of implementation schedule. Fed Regist. 2011;76:15282–90 [cited 2015 Apr 2]. https://www.federalregister.gov/articles/2011/03/21/2011-6585/new-performance-standards-for-salmonella-and-campylobacter-in-young-chicken-and-turkey-slaughter
- Kendall ME, Crim S, Fullerton K, Han PV, Cronquist AB, Shiferaw B, Travel-associated enteric infections diagnosed after return to the United States, Foodborne Diseases Active Surveillance Network (FoodNet), 2004–2009. Clin Infect Dis. 2012;54(Suppl 5):S480–7. DOIPubMedGoogle Scholar
- Hale CR, Scallan E, Cronquist AB, Dunn J, Smith K, Robinson T, Estimates of enteric illness attributable to contact with animals and their environments in the United States. Clin Infect Dis. 2012;54(Suppl 5):S472–9. DOIPubMedGoogle Scholar
- Gu W, Vieira AR, Hoekstra RM, Griffin PM, Cole D. Use of random forest to estimate population attributable fractions from a case–control study of Salmonella enterica serotype Enteritidis infections. Epidemiol Infect. 2015;Feb:1–9; Epub ahead of print .PubMedGoogle Scholar
- Varma JK, Samuel MC, Marcus R, Hoekstra RM, Medus C, Segler S, Listeria monocytogenes infection from foods prepared in a commercial establishment: a case–control study of potential sources of sporadic illness in the United States. Clin Infect Dis. 2007;44:521–8. DOIPubMedGoogle Scholar
- McCollum JT, Cronquist AB, Silk BJ, Jackson KA, O’Connor KA, Cosgrove S, Multistate outbreak of listeriosis associated with cantaloupe. N Engl J Med. 2013;369:944–53. DOIPubMedGoogle Scholar
- Shiferaw B, Verrill L, Booth H, Zansky SM, Norton DM, Crim S, Sex-based differences in food consumption: Foodborne Diseases Active Surveillance Network (FoodNet) population survey, 2006–2007. Clin Infect Dis. 2012;54(Suppl 5):S453–7. DOIPubMedGoogle Scholar
- Sheth AN, Hoekstra M, Patel N, Ewald G, Lord C, Clarke C, A national outbreak of Salmonella serotype Tennessee infections from contaminated peanut butter: a new food vehicle for salmonellosis in the United States. Clin Infect Dis. 2011;53:356–62. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Outbreak of Salmonella Heidelberg infections linked to a single poultry producer—13 states, 2012–2013. MMWR Morb Mortal Wkly Rep. 2013;62:553–6 .PubMedGoogle Scholar
- Abrams JY, Maddox RA, Harvey AR, Schonberger LB, Belay ED. Travel history, hunting, and venison consumption related to prion disease exposure, 2006–2007 FoodNet Population Survey. J Am Diet Assoc. 2011;111:858–63. DOIPubMedGoogle Scholar
- Voetsch AC, Kennedy MH, Keene WE, Smith KE, Rabatsky-Ehr T, Zansky S, Risk factors for sporadic Shiga toxin–producing Escherichia coli O157 infections in FoodNet sites, 1999–2000. Epidemiol Infect. 2007;135:993–1000. DOIPubMedGoogle Scholar
- Hurd S, Patrick M, Hatch J, Clogher P, Wymore K, Cronquist AB, Clinical laboratory practices for the isolation and identification of Campylobacter in Foodborne Diseases Active Surveillance Network (FoodNet) sites: baseline information for understanding changes in surveillance data. Clin Infect Dis. 2012;54(Suppl 5):S440–5. DOIPubMedGoogle Scholar
- Hoefer D, Hurd S, Medus C, Cronquist A, Hanna S, Hatch J, Laboratory practices for the identification of Shiga toxin-–roducing Escherichia coli in the United States, FoodNet sites, 2007. Foodborne Pathog Dis. 2011;8:555–60. DOIPubMedGoogle Scholar
- Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Foodborne illness acquired in the United States—major pathogens. Emerg Infect Dis. 2011;17:7–15. DOIPubMedGoogle Scholar
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–25. DOIPubMedGoogle Scholar
- Scallan E, Griffin PM, Angulo FJ, Tauxe RV, Hoekstra RM. Foodborne illness acquired in the United States—unspecified agents. Emerg Infect Dis. 2011;17:16–22. DOIPubMedGoogle Scholar
- Painter JA, Hoekstra RM, Ayers T, Tauxe RV, Braden CR, Angulo FJ, Attribution of foodborne illnesses, hospitalizations, and deaths to food commodities by using outbreak data, United States, 1998–2008. Emerg Infect Dis. 2013;19:407–15. DOIPubMedGoogle Scholar
- Kassenborg HD, Smith KE, Vugia DJ, Rabatsky-Ehr T, Bates MR, Carter MA, Fluoroquinolone-resistant Campylobacter infections: eating poultry outside of the home and foreign travel are risk factors. Clin Infect Dis. 2004;38(Suppl 3):S279–84. DOIPubMedGoogle Scholar
- Nelson JM, Chiller TM, Powers JH, Angulo FJ. Fluoroquinolone-resistant Campylobacter species and the withdrawal of fluoroquinolones from use in poultry: a public health success story. Clin Infect Dis. 2007;44:977–80. DOIPubMedGoogle Scholar
- Krueger AL, Greene SA, Barzilay EJ, Henao O, Vugia D, Hanna S, Clinical outcomes of nalidixic acid, ceftriaxone, and multidrug-resistant nontyphoidal salmonella infections compared with pansusceptible infections in FoodNet sites, 2006–2008. Foodborne Pathog Dis. 2014;11:335–41. DOIPubMedGoogle Scholar
- Shiferaw B, Solghan S, Palmer A, Joyce K, Barzilay EJ, Krueger A, Antimicrobial susceptibility patterns of Shigella isolates in Foodborne Diseases Active Surveillance Network (FoodNet) sites, 2000–2010. Clin Infect Dis. 2012;54(Suppl 5):S458–63. DOIPubMedGoogle Scholar
- O'Donnell AT, Vieira AR, Huang JY, Whichard J, Cole D, Karp BE. Quinolone-resistant Salmonella enterica serotype Enteritidis infections associated with international travel. Clin Infect Dis. 2014;59:e139–41. DOIPubMedGoogle Scholar
- Bemis K, Marcus R, Hadler JL. Socioeconomic status and campylobacteriosis, Connecticut, USA, 1999–2009. Emerg Infect Dis. 2014;20:1240–2 .PubMedGoogle Scholar
- Mody RK, Meyer S, Trees E, White PL, Nguyen T, Sowadsky R, Outbreak of Salmonella enterica serotype I 4,5,12:i:- infections: the challenges of hypothesis generation and microwave cooking. Epidemiol Infect. 2014;142:1050–60 . DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach—United States, September 2006. MMWR Morb Mortal Wkly Rep. 2006;55:1045–6 .PubMedGoogle Scholar
- Scallan E, Majowicz S, Hall G, Banerjee A, Bowman C, Daly L, Prevalence of diarrhoea in the community in Australia, Canada, Ireland, and the United States. Int J Epidemiol. 2005;34:454–60. DOIPubMedGoogle Scholar
- Dalton CB, Mintz ED, Wells JG, Bopp CA, Tauxe RV. Outbreaks of enterotoxigenic Escherichia coli infection in American adults: a clinical and epidemiologic profile. Epidemiol Infect. 1999;123:9–16. DOIPubMedGoogle Scholar
- Patrick ME, Mahon BE, Zansky SM, Hurd S, Scallan E. Riding in shopping carts and exposure to raw meat and poultry products: prevalence of, and factors associated with, this risk factor for Salmonella and Campylobacter infection in children younger than 3 years. J Food Prot. 2010;73:1097–100 .PubMedGoogle Scholar
- Scallan E, Jones T, Cronquist A, Thomas S, Frenzen P, Hoefer D, Factors associated with seeking medical care and submitting a stool sample in estimating the burden of foodborne illness. Foodborne Pathog Dis. 2006;3:432–8. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2013 [cited 2015 Apr 2]. http://www.cdc.gov/drugresistance/threat-report-2013/index.html
- President’s Council of Advisors on Science and Technology. National strategy for combating antibiotic-resistant bacteria. Sep 2014 [cited 2015 Apr 2]. https://www.whitehouse.gov/sites/default/files/docs/carb_national_strategy.pdf
Figures
Table
Cite This ArticleTable of Contents – Volume 21, Number 9—September 2015
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Olga L. Henao, Centers for Disease Control and Prevention, 1600 Clifton Rd NE, Mailstop C09, Atlanta, GA 30329-4027, USA
Top