Volume 9, Number 12—December 2003
Research
The Rabbit as a New Reservoir Host of Enterohemorrhagic Escherichia coli
Abstract
We investigated the prevalence of enterohemorrhagic Escherichia coli (EHEC) in rabbits acquired from two commercial vendors and a local petting zoo. Fecal samples from 34 Dutch Belted (DB) and 15 New Zealand White (NZW) rabbits were cultured; and isolates were biotyped, serotyped, tested by polymerase chain reaction (PCR), and genotyped by repetitive-element sequence–based PCR (Rep-PCR). Seven (25%) of 28 DB rabbits acquired from one commercial source were positive for EHEC, including O153:H- and O153:H7. One (9%) of 11 NZW rabbits from the same source was positive for eae-, stx1+ O153 strains. In contrast, six DB rabbits from another commercial source and four rabbits from a petting zoo were negative for EHEC. Rep-PCR demonstrated that the O153 EHEC and O145 enteropathogenic E. coli were two distinct clones. Our study indicates that rabbits are a new reservoir host of EHEC that may pose a zoonotic risk for humans.
Escherichia coli O157:H7 is a leading foodborne enteric pathogen associated with human illness, including hemorrhagic colitis and hemolytic uremic syndrome (HUS), the leading cause of acute renal failure in children (1). Similarly, non-O157 enterohemorrhagic E. coli (EHEC) serotypes have been implicated in outbreaks of disease worldwide and are currently considered emerging pathogens by the World Health Organization (WHO) (2). Most EHEC infections in humans are foodborne, and the source of infection is an animal reservoir. Cattle and other ruminants are considered major reservoirs hosts of EHEC (3). Recent reports have also emphasized that farm animals and their environment pose a zoonotic risk for humans, based on outbreaks of E. coli O157:H7 infection among farm visitors (4).
Identifying bacterial pathogens in natural hosts is important because they constitute potential reservoirs for zoonotic transmission (5). We recently described an outbreak of hemorrhagic diarrhea and hemolytic uremic syndrome (HUS) in Dutch Belted (DB) rabbits naturally infected with EHEC O153:H- (6). In the current study, we investigated the prevalence of EHEC in laboratory rabbits acquired from two commercial vendors and in rabbits from a local petting zoo to assess their potential as reservoir hosts.
Rabbits
Fecal samples were collected from 34 DB and 15 New Zealand White (NZW) Pasturella multocida–free laboratory rabbits acquired at various times from one commercial source. Fecal samples were also collected from six DB rabbits that were acquired from a second vendor and from four rabbits of various breeds that belonged to a local petting zoo.
Bacterial Culture and Isolation
Fecal pellets were homogenized in tryptic soy broth (Remel, Lenexa, KS) and incubated at 37°C overnight. Each sample was then plated on Rainbow Agar O157 (Biolog, Hayward, CA) and incubated at 37°C for approximately 36 hours. Bacterial colonies were selected on the basis of color. Pink and purple colonies were selected and restreaked on MacConkey or blood agar plates (Remel) or both. All the E. coli isolates were confirmed biochemically and characterized by using API 20E strips (Biomerieux Vitek, Hazelwood, MO). Selected E. coli organisms were serotyped at the Pennsylvania State University E. coli Reference Center (ECRC).
DNA Extraction and Polymerase Chain Reaction (PCR)
Genomic DNA was extracted from bacteria by using InstaGene Matrix (Bio-Rad Laboratories, Hercules, CA). PCR primers and conditions to detect eae, stx1B, and stx2A and variants have been described previously (6).
Southern Blot Analysis
Southern blot analysis was performed by using a Shiga toxin 1B (stx1B) probe generated by PCR amplification of EHEC O153:H- DNA to confirm stx1B in EHEC strains isolated from rabbits. Fifteen microliters of amplicon underwent electrophroresis through a 1.3% agarose gel and was transferred onto a Hybond N nylon membrane, as outlined by the manufacturer (Amersham, Piscataway, NJ). DNA was then cross-linked using the UV Stratalinker 1800 (Stratagene, La Jolla, CA). The fixed DNA was subsequently hybridized with the stx1B probe. The probe was labeled with horseradish peroxidase, exposed in the presence of luminol to Hyperfilm-ECL as outlined by the manufacturer (Amersham).
High-Resolution Genotyping by Repetitive-Element Sequence-Based PCR (Rep-PCR)
To perform Rep-PCR chromosomal profiling, genomic DNA was extracted from bacteria by using High Pure PCR Template Preparation Kit (Roche Diagnostics GmbH, Mannheim, Germany) and quantified by using a GeneQuant pro (Biochrom Ltd., Cambridge, UK) spectrophotometer. Two primer pairs were used for the amplification reactions (7): REP1R-I (5′-IIIICGICGICATCIGGC-3′), REP2-I (5′-ICGICTTATCIGGCCTAC-3′); and ERIC1R (5′-ATGTAAGCTCCTGGGGATTCAC-3′), ERIC2 (5′-AAGTAAGTGACTGGGGTGAGCG-3′). PCR reaction mixtures were prepared by using puReTaq Ready-To-Go PCR Beads (Amersham) containing 50 pmol of each primer and 1 μL (100 ng) of DNA to a total volume of 25 μL. Rep-PCR reactions were performed by using a Techne Genius (Techne Inc., Princeton, NJ) thermal cycler with the following conditions: for the REP primers, initial denaturation (94°C, 7 min), followed by 30 cycles of denaturation (94°C, 30 s), annealing (40°C, 1 min), and extension (65°C, 8 min). A final extension (65°C, 16 min) completed the cycling protocol. For the ERIC primers, initial denaturation (95°C, 5 min) was followed by 35 cycles of denaturation (92°C, 45 s), annealing (51°C, 1 min), and extension (70°C, 10 min). A final extension (70°C, 20 min) completed the cycling protocol. PCR amplicons were visualized after electrophoresis in a 3% agarose gel and staining with ethidium bromide.
Bacterial Cultures
Rabbit fecal samples cultured on Rainbow agar O157 yielded E. coli colonies that ranged in color from pink to purple. No black colonies consistent with E. coli O157 were observed (8).
Biochemical and molecular characterization of the rabbit E. coli strains is summarized in the Table. Most O153 strains were negative for rhamnose and sucrose. Some colonies of the O153:H- EHEC strain isolated from the first outbreak of HUS in DB rabbits (6) and a strain of unknown O serotype that showed autoagglutination were sorbitol negative like most O157:H7 EHEC organisms isolated from humans. In addition, two O145:H- EPEC were sorbitol negative.
Prevalence of EHEC Strains
Seven (25%) of 28 DB rabbits acquired from one commercial source were positive for eae+, stx1+ E. coli, including serotypes O153:H- and O153:H7. One (14%) of the 7 rabbits was infected with an EHEC of unknown O serotype that showed autoagglutination (DB 02-177, isolate 03-192) and was co-infected with enteropathogenic E. coli (EPEC) O145:H-. Eleven (39%) of the 28 DB rabbits were positive for EPEC O145H- or O145:H7. In addition, 1 (9%) of 11 NZW rabbits from the same source was positive for eae-, stx1+ O153:H-, and O153:H7 E. coli. All the E. coli strains were negative for stx2 and variants. In contrast, six DB rabbits from another commercial source and all four rabbits from the petting zoo were negative for EHEC and EPEC. Almost all of the eae-, stx1- E. coli belonged to serotype O7:H7.
Southern Blot Analysis
The presence of the stx1B amplicon was confirmed in O153:H- and O153:H7 E. coli. The O145:H- and O145:H7 isolates were confirmed to be negative for stx1B (Figure 1).
High-Resolution Genotyping by Rep-PCR
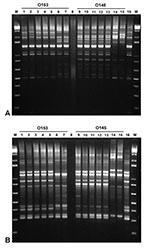
Figure 2 shows the results obtained by Rep-PCR by using two different sets of primers, REP and ERIC. All the rabbit O153:H– and O153:H7 Stx–producing E. coli organisms tested, including eae- isolates from a NZW rabbit, produced identical amplification patterns that differed from those produced by two other O153:H- EHEC strains isolated from humans. Similarly, all the rabbit O145:H- and O145:H7 EPEC isolates produced identical amplification patterns that differed from those produced by a human and a bovine O145:H- EHEC. The EHEC isolated from the rabbit infected with EPEC O145:H- had an amplification pattern similar to that produced by the O153 strains.
In 2001, approximately 2 million rabbits were used as a food source (9). In addition, in 2000, laboratories used >250,000 rabbits, and the domestic rabbit population in the United States was estimated at 9 million (9). Also, the rabbit is becoming an increasingly popular pet in U.S. households. In this study, we determined that 25% of the DB and 9% of the NZW rabbits from one commercial source of laboratory rabbits harbored Stx-producing E. coli O153:H- or O153:H7 in their feces. These findings raise concerns about the zoonotic risk for humans, given that rabbits are common companion animals, are used for biomedical research, and are an agricultural food source. DB rabbits are very popular in pet stores, are commonly used in rabbit shows, and are the second most common rabbit breed used in research after NZW rabbits (9). In addition, NZW rabbits are commonly used as a meat source in the United States. More studies are needed to assess the prevalence of these strains and other EHEC in agricultural, pet, and wild rabbit populations. Wild rabbits were also identified as vectors of a Stx-producing O157 E. coli strain that was isolated from cattle in an outbreak of hemorrhagic diarrhea and HUS involving visitors to a zoo (10). Our finding that most O153:H- and O153:H7 EHEC were unable to ferment rhamnose and sucrose indicates that these biochemical markers may be useful for the detection of these strains. For example, the inability to ferment rhamnose by some human EHEC strains has been applied to the development of selective isolation media (11). Rhamnose-negative rabbit EPEC strains appear to be highly pathogenic (12).
Cattle are considered the primary reservoir host of O157 and non-O157 EHEC (13). Human infections have been linked to the presence of these bacteria in undercooked ground beef. In a recent study, the prevalence of non-O157 EHEC strains on beef carcasses was >50%. Some isolates in that study also belonged to serotypes O153 and O145 (13). In addition, a study investigating the prevalence of non-O157 EHEC from human diarrheal samples in the United States demonstrated that non-O157 serotypes are at least as prevalent as serotype O157 (14). Serotype O145 has been isolated from diarrheic children in the United States and is a common non-O157 EHEC serotype isolated from HUS case-patients in Europe (15,16). Indeed, non-O157 serotypes are a leading cause of HUS in Germany (17).
Co-infection of EHEC and EPEC was also identified in one rabbit in this study. The EHEC and EPEC strains isolated from this rabbit appeared to belong to different serotypes. The high prevalence of these E. coli strains in rabbits and the occurrence of co-infection suggest that in vivo transduction of EPEC by Stx-encoding bacteriophages from EHEC may naturally occur in this host (18). By performing Southern blot analysis, we confirmed the presence of stx1B in the O153:H- and O153:H7 isolates, including the strain isolated from the rabbit infected with EPEC. In addition, by performing Rep-PCR analysis, we demonstrated that selected rabbit O153 EHEC and O145 EPEC isolates of the same O serotype represented two distinct clones. This apparent clonal nature of the isolates suggests transmission of E. coli between rabbits. Because these rabbit E. coli strains appeared to represent distinct clonal groups by Rep-PCR fingerprinting, their phylogenetic relatedness to other strains was subsequently investigated by using multilocus sequence typing. EHEC O153 and EPEC O145 belong to the EHEC 2 and EPEC 2 groups, respectively (T. S. Whittam, unpub. data). Rabbits may have become infected with these EHEC and EPEC strains from hay contaminated with cattle feces. Alternatively, rabbit-adapted E. coli strains may have been transduced by Stx-encoding bacteriophages from EHEC strains that transiently infected this rabbit colony.
Previous experiments in NZW rabbits, in which purified Stx1 was infused intravenously, postulated that renal lesions did not develop in rabbits and that the rabbit model failed to replicate human HUS (19,20). We recently reported that glomerulonephritis, tubular lesions, and renal glomerular thrombotic microangiopathy, the hallmark of HUS, developed in DB rabbits naturally infected with EHEC (6). In the present study, however, HUS did not develop in most rabbits colonized with EHEC. This result is consistent with human studies in which asymptomatic infection has been reported in household contacts of children with HUS and with studies showing that previous infection and frequent reexposure to E. coli O157:H7 may confer some protection against symptomatic illness (4,21).
In summary, our findings indicate that rabbits are a newly recognized reservoir host of EHEC that poses a zoonotic risk to humans. These findings also provide an opportunity to develop a rabbit model to study the pathogenesis of EHEC-induced disease and HUS in a naturally susceptible reservoir host.
Dr. García is a postdoctoral fellow in the Division of Comparative Medicine at the Massachusetts Institute of Technology (MIT). His research interests include microbial pathogenesis and animal models of infection and neoplasia.
Dr. Fox is professor and director of the Division of Comparative Medicine and professor in the Biological Engineering Division at MIT. Dr. Fox’s research interests are centered on microbial pathogenesis and the role of chronic inflammation in cancer.
Acknowledgments
We thank David B. Schauer for critical review of the manuscript.
This study was supported by NIH grant T32-RR07036.
References
- Hughes AK, Ergonal Z, Stricklett PK, Kohan DE. Molecular basis for high renal cell sensitivity to the cytotoxic effects of shigatoxin-1: upregulation of globotriaosylceramide expression. J Am Soc Nephrol. 2002;13:2239–45. DOIPubMedGoogle Scholar
- World Health Organization. Zoonotic non-O157 shiga toxin-producing Escherichia coli (STEC). Report of a WHO scientific working group meeting. Berlin, Germany: Department of Communicable Disease Surveillance and Response, The Organization; 1998.
- Rabatsky-Ehr T, Dingman D, Marcus R, Howard R, Kinney A, Mshar P. Deer meat as the source for a sporadic case of Escherichia coli O157:H7 infection, Connecticut. Emerg Infect Dis. 2002;8:525–7.PubMedGoogle Scholar
- Crump JA, Sulka AC, Langer AJ, Schaben C, Crielly AS, Gage R, An outbreak of Escherichia coli O157:H7 infections among visitors to a dairy farm. N Engl J Med. 2002;347:555–60. DOIPubMedGoogle Scholar
- Haydon DT, Cleaveland S, Taylor LH, Laurenson MK. Identifying reservoirs of infection: a conceptual and practical challenge. Emerg Infect Dis. 2002;8:1468–73.PubMedGoogle Scholar
- García A, Marini RP, Feng Y, Vitsky A, Knox KA, Taylor NS, A naturally occurring rabbit model of enterohemorrhagic Escherichia coli-induced disease. J Infect Dis. 2002;186:1682–6. DOIPubMedGoogle Scholar
- Versalovic J, Kapur V, Koeuth T, Mazurek GH, Whittam TS, Musser JM, DNA fingerprinting of pathogenic bacteria by fluorophore-enhanced repetitive sequence-based polymerase chain reaction. Arch Pathol Lab Med. 1995;119:23–9.PubMedGoogle Scholar
- Bettelheim KA. Studies of Escherichia coli cultured on Rainbow Agar O157 with particular reference to enterohaemorrhagic Escherichia coli (EHEC). Microbiol Immunol. 1998;42:265–9.PubMedGoogle Scholar
- Grannis JUS. rabbit industry profile: USDA, APHIS, VS. Fort Collins (CO): Centers for Epidemiology and Animal Health, Center for Emerging Issues; 2002.
- Pritchard GC, Williamson S, Carson T, Bailey JR, Warner L, Willshaw G, Wild rabbits—a novel vector for verocytotoxigenic Escherichia coli O157. Vet Rec. 2001;149:567.PubMedGoogle Scholar
- Hiramatsu R, Matsumoto M, Miwa Y, Suzuki Y, Saito M, Miyazaki Y. Characterization of Shiga toxin–producing Escherichia coli O26 strains and establishment of selective isolation media for these strains. J Clin Microbiol. 2002;40:922–5. DOIPubMedGoogle Scholar
- Camguilhem R, Milon A. Biotypes and O serogroups of Escherichia coli involved in intestinal infections of weaned rabbits: clues to diagnosis of pathogenic strains. J Clin Microbiol. 1989;27:743–7.PubMedGoogle Scholar
- Arthur TM, Barkocy-Gallagher GA, Rivera-Betancourt M, Koohmaraie M. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli on carcasses in commercial beef cattle processing plants. Appl Environ Microbiol. 2002;68:4847–52. DOIPubMedGoogle Scholar
- Fey PD, Wickert RS, Rupp ME, Safranek TJ, Hinrichs SH. Prevalence of non-O157 Shiga toxin–producing Escherichia coli in diarrheal stool samples from Nebraska. Emerg Infect Dis. 2000;6:530–3. DOIPubMedGoogle Scholar
- Bokete TN, Whittam TS, Wilson RA, Clausen CR, O’Callahan CM, Moseley SL, Genetic and phenotypic analysis of Escherichia coli with enteropathogenic characteristics isolated from Seattle children. J Infect Dis. 1997;175:1382–9. DOIPubMedGoogle Scholar
- Eklund M, Scheutz F, Siitonen A. Clinical isolates of non-O157 Shiga toxin-producing Escherichia coli: serotypes, virulence characteristics, and molecular profiles of strains of the same serotype. J Clin Microbiol. 2001;39:2829–34. DOIPubMedGoogle Scholar
- Verweyen HM, Karch H, Allerberger F, Zimmerhackl LB. Enterohemorrhagic Escherichia coli (EHEC) in pediatric hemolytic-uremic syndrome: a prospective study in Germany and Austria. Infection. 1999;27:341–7. DOIPubMedGoogle Scholar
- Acheson DW, Reidl J, Zhang X, Keusch GT, Mekalanos JJ, Waldor MK. In vivo transduction with shiga toxin 1-encoding phage. Infect Immun. 1998;66:4496–8.PubMedGoogle Scholar
- Zoja C, Corna D, Farina C, Sacchi G, Lingwood C, Doyle MP, Verotoxin glycolipid receptors determine the localization of microangiopathic process in rabbits given verotoxin-1. J Lab Clin Med. 1992;120:229–38.PubMedGoogle Scholar
- Richardson SE, Rotman TA, Jay V, Smith CR, Becker LE, Petric M, Experimental verocytotoxemia in rabbits. Infect Immun. 1992;60:4154–67.PubMedGoogle Scholar
- Ludwig K, Sarkim V, Bitzan M, Karmali MA, Bobrowski C, Ruder H, Shiga toxin-producing Escherichia coli infection and antibodies against Stx2 and Stx1 in household contacts of children with enteropathic hemolytic-uremic syndrome. J Clin Microbiol. 2002;40:1773–82. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 9, Number 12—December 2003
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Alexis García, Division of Comparative Medicine, Massachusetts Institute of Technology, 77 Massachusetts Avenue, 16-820, Cambridge, MA 02139, USA; phone 617-253-1757, fax: 617-258-5708
Top