Volume 19, Number 8—August 2013
Dispatch
Recombinant Coxsackievirus A2 and Deaths of Children, Hong Kong, 2012
Figure 2
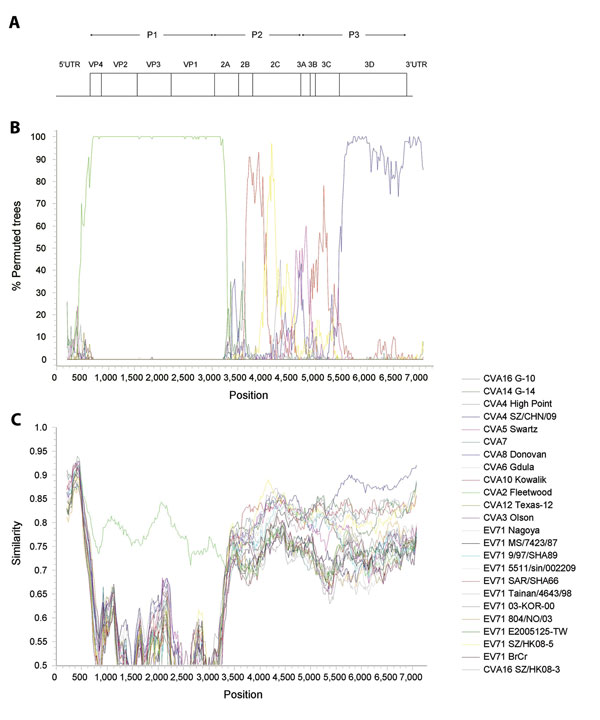
Figure 2. . Recombination analysis of complete genome of coxsackievirus A2 (CVA2), Hong Kong, 2012. A) Genome organization. B) Bootscanning and C) similarity plot analyses were conducted by using Simplot version 3.5.1 (http://sray.med.som.jhmi.edu/SCRoftware/simplot/) (Kimura distance model; window size, 400 bp; step, 20 bp) on a gapless nucleotide alignment generated with ClustalX 2.0 (8) with the genome sequence of CVA2 strain 430895 as the query sequence. In panel B, green line indicates CVA2 prototype strain Fleetwood, red line indicates EV71 strain SAR/SHA66 of subgenotype B3, yellow line indicates EV71 strain SZ/HK08–5 of subgenotype C4, and blue line indicates CVA4 strain SZ/CHN/09. P1, capsid protein 1, P2, nonstructural protein 2; P3, nonstructural protein 3; UTR, untranslated region; VP, viral protein.
References
- Pallansch M. Enteroviruses: polioviruses, coxsackieviruses, echoviruses and newer enteroviruses. In: Knipe DM, Howley PM, editors. Fields virology, 5th ed. Philadelphia: Wolters Kluwer Health/Lippincott Williams and Wilkins; 2007. p. 840–93.
- Wong SS, Yip CC, Lau SK, Yuen KY. Human enterovirus 71 and hand, foot and mouth disease. Epidemiol Infect. 2010;138:1071–89. DOIPubMedGoogle Scholar
- Lau SK, Yip CC, Lung DC, Lee P, Que TL, Lau YL, Detection of human rhinovirus C in fecal samples of children with gastroenteritis. J Clin Virol. 2012;53:290–6. DOIPubMedGoogle Scholar
- Yip CC, Lau SK, Woo PC, Chan KH, Yuen KY. Complete genome sequence of a coxsackievirus A22 strain in Hong Kong reveals a natural intratypic recombination event. J Virol. 2011;85:12098–9. DOIPubMedGoogle Scholar
- Yip CC, Lau SK, Zhou B, Zhang MX, Tsoi HW, Chan KH, Emergence of enterovirus 71 “double-recombinant” strains belonging to a novel genotype D originating from southern China: first evidence for combination of intratypic and intertypic recombination events in EV71. Arch Virol. 2010;155:1413–24 and. DOIPubMedGoogle Scholar
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. DOIPubMedGoogle Scholar
- Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McInerney JO. Assessment of methods of amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. DOIPubMedGoogle Scholar
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. DOIPubMedGoogle Scholar
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Full-length human immunodeficiency virus type 1 genomes from subtype C–infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–60 .PubMedGoogle Scholar
- Lukashev AN. Role of recombination in evolution of enteroviruses. Rev Med Virol. 2005;15:157–67 . DOIPubMedGoogle Scholar
- McWilliam Leitch EC, Cabrerizo M, Cardosa J, Harvala H, Ivanova OE, Koike S, The association of recombination events in the founding and emergence of subgenogroup evolutionary lineages of human enterovirus 71. J Virol. 2012;86:2676–85 . DOIPubMedGoogle Scholar