Volume 11, Number 1—January 2005
Dispatch
G, N, and P Gene-based Analysis of Chandipura Viruses, India
Figure 5
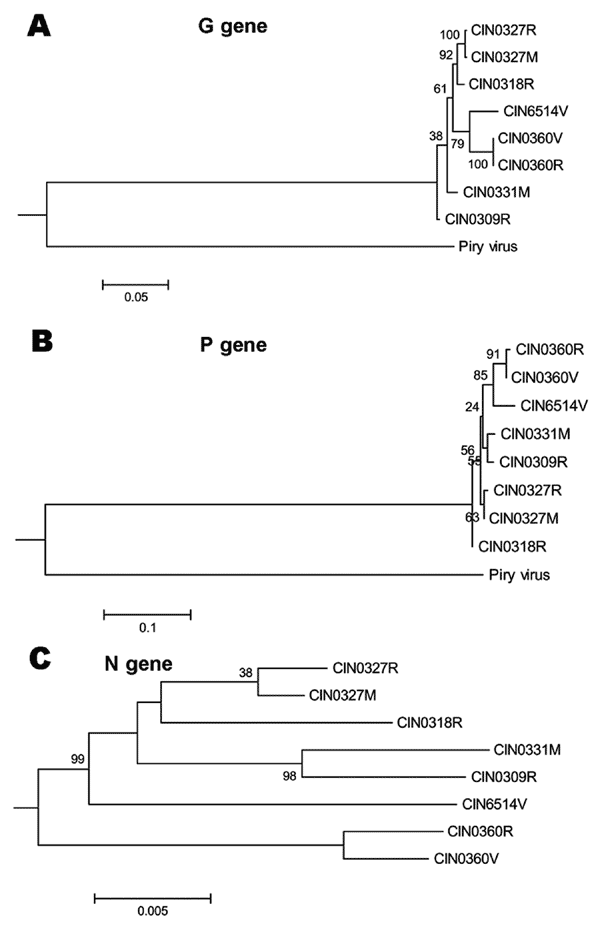
Figure 5. Phylogenetic analyses of complete G gene (A), P gene (B), and N gene (C) of Chandipura virus isolates. For details on isolates, see Table 2. Percent bootstrap support is indicated by the values at each node. For G and P gene-based analyses, Piry virus (GenBank accession no. D26175) was used as an outgroup. For N gene, an unrooted tree was constructed because the sequence for Piry virus was not available.
Page created: April 15, 2011
Page updated: April 15, 2011
Page reviewed: April 15, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.