Volume 11, Number 2—February 2005
Dispatch
Diagnostic System for Rapid and Sensitive Differential Detection of Pathogens
Figure 2
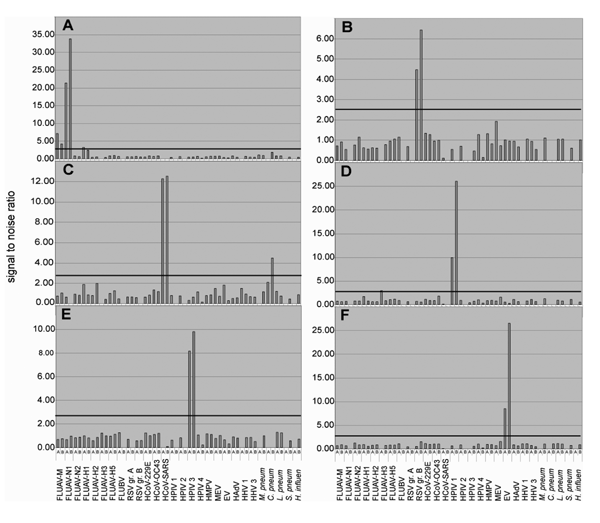
Figure 2. . Analysis of clinical specimens. RNA extracts from clinical specimens containing known pathogens were reverse transcribed into cDNA (Superscript RT system, Invitrogen, Carlsbad, CA; 20-µL volume). Five microliters of the reaction were subjected to Mass Tag PCR by using primers coupled to Masscode tags (Qiagen Masscode technology, Qiagen, Hilden, Germany). Detection of (A) influenza virus A (H1N1), (B) respiratory syncytial virus (RSV) group B, (C) human coronavirus SARS (HCoV-SARS), (D) human parainfluenza virus (HPIV) types 1 and (E) 3, and (F) enterovirus (EV) by using a 30-plex assay, including 60 primers targeting influenza A virus matrix gene (FLUAV-M), and for typing N1, N2, H1, H2, H3, and H5 sequences, as well as influenza B virus (FLUBV), RSV groups A and B, HCoV-229E, -OC43, and -SARS, HPIV types 1, 2, 3, and 4 (groups A and B combined; 4 primers), human metapneumovirus (HMPV, 4 primers), measles virus (MEV), EV (degenerate primer pair targeting all serogroups), human adenoviruses (HAdV, degenerate primer pair targeting all serogroups), human herpesvirus 1 (HHV-1, herpes simplex virus), human herpesvirus 3 (HHV-3; varicella-zoster virus), Mycoplasma pneumoniae, Chlamydia pneumoniae, Legionella pneumophila, Streptococcus pneumoniae, Haemophilus influenzae. The bar indicates an arbitrary cut-off threshold of 2.7 (4 times average background determined with random human DNA).
1These authors contributed equally to this study.