Volume 12, Number 3—March 2006
Synopsis
Web-based Surveillance and Global Salmonella Distribution, 2000–2002
Abstract
Salmonellae are a common cause of foodborne disease worldwide. The World Health Organization (WHO) supports international foodborne disease surveillance through WHO Global Salm-Surv and other activities. WHO Global Salm-Surv members annually report the 15 most frequently isolated Salmonella serotypes to a Web-based country databank. We describe the global distribution of reported Salmonella serotypes from human and nonhuman sources from 2000 to 2002. Among human isolates, S. Enteritidis was the most common serotype, accounting for 65% of all isolates. Among nonhuman isolates, although no serotype predominated, Salmonella enterica serovar Typhimurium was reported most frequently. Several serotypes were reported from only 1 region of the world. The WHO Global Salm-Surv country databank is a valuable public health resource; it is a publicly accessible, Web-based tool that can be used by health professionals to explore hypotheses related to the sources and distribution of salmonellae worldwide.
Foodborne diseases are among the most serious health problems affecting public health and development worldwide (1). Industrialization, mass food production, decreasing trade barriers, and human migration have disseminated and increased the incidence and severity of foodborne diseases worldwide (2–4).
Salmonellae are among the most common bacterial foodborne pathogens worldwide (4). They cause an estimated 1.4 million cases of foodborne disease each year in the United States alone (5). Salmonella serotyping is a surveillance tool that detects widespread outbreaks, identifies outbreak sources, monitors trends over time, and attributes human disease to various foods and animals (6). Such surveillance is needed to help prevent foodborne disease outbreaks and raise awareness among health authorities, food producers, food regulators, and consumers (7).
A 1997 survey of national reference laboratories showed that only 69 (66%) of 104 responding countries conducted routine Salmonella serotyping for public health surveillance (8). Consequently, the World Health Organization (WHO), the US Centers for Disease Control and Prevention, and the Danish Veterinary Laboratory (now the Danish Institute for Food and Veterinary Research) founded WHO Global Salm-Surv in 2000. Its mission is to promote integrated, laboratory-based surveillance and foster collaboration among human health, veterinary, and food-related disciplines to enhance the capacity to detect, respond, and prevent foodborne diseases (9). By November 2005, WHO Global Salm-Surv had >800 members from 142 countries. A key component of this program is the Web-based country databank, to which member countries annually submit their 15 most frequently isolated Salmonella serotypes. This program is the only foodborne disease surveillance network that is global in scope and surveys all aspects of the food chain, from animal feed to humans. Data are updated annually and are publicly accessible for members and the scientific community to review (http://www.who.int/salmsurv). We describe the global distribution of Salmonella serotypes from human and nonhuman sources reported to the WHO Global Salm-Surv country databank from 2000 to 2002 and explore how the databank may become a valuable public health resource for foodborne disease surveillance.
WHO Global Salm-Surv has conducted annual regional training courses for national reference laboratories since 1999 and has managed an external laboratory quality assurance program since 2000 to facilitate a standard approach to isolating and serotyping salmonellae (10). National reference laboratories can become WHO Global Salm-Surv members and share Salmonella serotype data with other members through the country databank. The country databank is a Web-enabled Oracle database that is password protected for data entry and accessible for public viewing at http://www.who.int/salmsurv. Each year, a designated national reference laboratory representative enters into the country databank the number of Salmonella isolates serotyped from human, animal, food, environmental, and feed sources and the 15 most frequently identified serotypes.
Descriptive analysis was conducted by using Microsoft Excel (Microsoft Corp., Redmond, WA, USA) on data from all countries that submitted data for 2000, 2001, or 2002 as of June 2004. Analyses for trends over time were conducted on data from 2000 to 2002. More detailed analyses, including ranking of serotypes, comparison of human to nonhuman isolates, and regional comparisons are presented for 2002 data only, the year in which the most countries participated.
Before 2001, nonhuman isolates were grouped together. Since 2001, countries have been able to submit food, animal, environmental, and feed data separately. For comparison purposes, all nonhuman data were combined in this analysis.
Data were grouped into regions approximately corresponding to 6 geopolitical continents: Africa, Asia, Latin America and the Caribbean, Europe, North America, and Oceania. To accommodate local epidemiologic characteristics, New Caledonia was incorporated into Asia, and Israel was incorporated into Europe. For years in which a single country contributed data for a region, regional data are not presented, but the data are included in the overall results. A region-specific serotype was defined as a serotype that, for each of the years of the study period, was among the 15 most commonly reported serotypes and for which >90% of the isolates were from that region.
Global
Forty-nine countries submitted data to the WHO Global Salm-Surv country databank from 2000 to 2002 (Table 1). Twenty countries reported both human and nonhuman results, 21 reported only human results, and 8 reported only nonhuman results. Reports of 376,856 human and 65,789 nonhuman Salmonella isolations were entered into the database during the 3-year period. North America and Europe accounted for 87.9% (389,134) of all reported isolates. The number of isolates reported to the country databank was stable during the study period; 113,782–137,329 human isolates and 16,506–25,761 nonhuman isolates were reported per year.
During the 3-year period, Salmonella enterica serovar Enteritidis was by far the most common serotype reported from human isolates globally. In 2002, it accounted for 65% of all isolates, followed by S. Typhimurium at 12% and S. Newport at 4%. Among nonhuman isolates, S. Typhimurium was the most commonly reported serotype in all 3 years, accounting for 17% of isolates in 2002. It was followed by S. Heidelberg (11%) and S. Enteritidis (9%) (Figure 1).
In 2002, 26 (84%) of the 31 countries that reported human serotype results ranked S. Enteritidis and S. Typhimurium in their 10 most common human serotypes (Table 2). Approximately half of the countries ranked S. Infantis and S. Typhi in their 10 most common serotypes, but only a fourth ranked S. Newport and S. Heidelberg in their top 10. The relative ranking of serotypes by the number of countries reporting them in their 15 most frequent serotypes remained stable over the study period (data not shown). However, the proportion of countries reporting each serotype varied. For example, from 2000 to 2001, more than two thirds of countries reported S. Agona, compared to 39% in 2002.
In 2002, a total of 5 serotypes were reported among the 15 most common human serotypes from all 6 regions of the world: S. Enteritidis, S. Typhimurium, S. Infantis, S. Montevideo, and S. Typhi. However, the proportion of isolates of each serotype varied greatly. In 2002, for example, S. Enteritidis represented 85% of isolates in Europe but only 9% in Oceania. In Latin America and the Caribbean, S. Typhi accounted for the greatest proportion of salmonellae (13%). In 2000 and 2001, S. Enteritidis, S. Typhimurium, S. Typhi, and S. Agona were reported from all 6 regions (data not shown).
S. Enteritidis, S. Typhimurium, and S. Typhi were ranked among the 15 most common human serotypes in all 6 regions throughout the 3-year study period. S. Agona, S. Infantis, S. Montevideo, S. Saintpaul, S. Hadar, S. Mbandaka, S. Newport, S. Thompson, S. Heidelberg, and S. Virchow were also widespread; they were reported from 4 to 6 of the regions from 2000 through 2002. Reporting of S. Montevideo increased from 4 regions in 2000 to all 6 regions in 2002. The reporting of S. Heidelberg increased from 3 to 5 regions in the same timeframe.
Regional
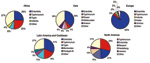
In Africa in 2002, S. Enteritidis and S. Typhimurium were each reported from approximately one fourth of isolates from humans (Figure 2). Among nonhuman sources (Figure 3), S. Anatum and S. Enteritidis constituted the largest proportion of isolates.
In Asia, from 2000 through 2002, Japan, Korea, and Thailand together reported S. Enteritidis as the most common human serotype. S. Weltevreden was the second most common serotype in 2000 and 2001 but dropped to fourth in 2002, when it was surpassed by S. Rissen and S. Typhimurium. In 2002, S. Enteritidis accounted for 38% of human isolates but only 7% of nonhuman isolates. S. Anatum, S. Rissen, and S. Stanley were the most common nonhuman serotypes in Asia.
In Europe in 2002, S. Enteritidis accounted for most salmonellae among human isolates. This trend was constant from 2000 to 2002; S. Enteritidis accounted for 79% to 84% of isolates, followed by S. Typhimurium in second place and S. Hadar, S. Virchow, and S. Infantis alternating in the third to fifth places among the 8 countries that submitted data during the 3 years. Among nonhuman isolates, heterogeneity was greater; S. Infantis, S. Enteritidis, and S. Typhimurium together accounted for 72% of salmonellae in 2002.
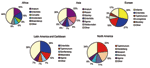
In 2002 in Latin America and the Caribbean, S. Enteritidis was the most common serotype among human and nonhuman isolates. S. Typhimurium, S. Typhi, S. Montevideo, and S. Paratyphi B were also commonly observed among human isolates and S. Typhimurium, S. Senftenberg, S. Mbandaka, and S. Agona, among nonhuman isolates. During the 3-year period of interest, S. Enteritidis, S. Typhimurium, and S. Typhi were the 3 most commonly isolated serotypes among humans in the 5 countries that reported data every year.
In North America in 2002, S. Typhimurium was more common than S. Enteritidis among human isolates. S. Newport and S. Heidelberg also accounted for a sizeable proportion of the isolates. Among nonhuman isolates, a corresponding pattern emerges; S. Typhimurium, S. Heidelberg, and S. Newport were most common. S. Enteritidis was not reported among the 10 most common nonhuman serotypes. The relative ranking of serotypes did not change in the 3-year period; S. Typhimurium was the most common serotype in humans and nonhuman isolates from 2000 to 2002. In Oceania in 2000, the only year in which >1 country reported data, S. Typhimurium accounted for 62% of human Salmonella isolates, followed by S. Virchow and S. Enteritidis.
Some serotypes were reported among the 15 most common serotypes in only 1 region during the 3-year period and therefore were classified as region-specific serotypes. Africa was the only region to report S. Brancaster among nonhuman isolates. Asia was the only region to report S. Rissen (human), S. Panama and S. Stanley (nonhuman), and S. Weltevreden (human and nonhuman). Europe was the only region to report S. Blockley, S. Kisangani, S. Kottbus, S. Ohio, and S. Stanleyville from human isolates and S. Indiana and S. Isangi from nonhuman isolates. Latin America and the Caribbean was the only region to report S. Bardo, S. Muenster, and S. Rubislaw among human isolates. North America was the only region to report S. Javiana (human) and S. Muenster (nonhuman).
S. Enteritidis is the most common Salmonella serotype in humans globally but especially in Europe, where it accounts for 85% of Salmonella cases, Asia (38%), and Latin America and the Caribbean (31%). The S. Enteritidis pandemic was first noted in the late 1980s and has been attributed to contaminated eggs (11). The proportion of Salmonella infections associated with this serotype seems to have increased over time. In 1995, 36% of salmonellae worldwide were S. Enteritidis, compared to 65% in 2002 (8).
S. Typhimurium has been 1 of the 2 most frequent serotypes in humans since 1990 (8). Since S. Enteritidis and S. Typhimurium are so common, additional subtyping methods, including phage typing, antimicrobial susceptibility testing, and pulsed-field gel electrophoresis (PFGE), are needed to identify clusters of infection from the same source. WHO Global Salm-Surv includes antimicrobial susceptibility testing training in all regional courses and has introduced phage typing in the Eastern European region course. The country databank could include data from such subtyping efforts. PFGE subtyping data are exchanged in North American between PulseNet USA and PulseNet Canada (12). PulseNet International is an affiliate member of WHO Global Salm-Surv, and the networks are coordinating their efforts to ensure synergy.
S. Typhi is a pathogen of concern in the developing world, especially Asia (13). However, in our analyses, S. Typhi was the ninth most frequent serotype in Asia in 2002. The Asian countries that contributed to the country databank did not include many of the developing countries in south-central and Southeast Asia, where S. Typhi is still highly prevalent. S. Typhi was the sixth most frequent serotype globally in 1995 and was decreasing in relative importance (8). That trend seems to have continued; S. Typhi ranked 14th globally in 2002. S. Typhi has no animal reservoir, which makes it susceptible to improvements in hygiene and sanitation seen in many regions of the world, such as Latin America and the Caribbean.
The distribution of nonhuman serotypes is more heterogeneous than that of human serotypes. The same serotypes appear among the top 5 in human and nonhuman sources, although in a different order. S. Enteritidis is only the third most common serotype among nonhuman sources. In 2002, it was not reported at all among the 10 most common nonhuman serotypes from North America. This finding partly reflects the capacity of S. Enteritidis to contaminate eggs in low numbers and the difficulty of isolating it from food or the environment. Moreover, in North America, few samples from eggs are submitted for routine testing. For example, in the United States, routine testing of eggs is not required, whereas routine testing for salmonellae is required of meat and poultry plants. As eggs are frequently used in foods that do not undergo heat treatment (e.g., pastries, homemade ice cream, and mayonnaise) and are widely distributed, this food contamination has a substantial effect on public health.
The country databank contains far fewer nonhuman than human serotypes, possibly because more participating laboratories are human national reference laboratories, fewer countries have formal nonhuman surveillance, and some countries may be less likely to share nonhuman data because of trade concerns. In 2001 and 2002, 15 of 22 countries reported nonhuman isolates by source. Food serotypes were reported from most countries (11 in 2001 and 12 in 2002), followed by animal serotypes (7 countries in 2001 and 10 in 2002). Most isolates serotyped were from animals (66%), followed by food (29%), feed (3%), and the environment (2%). The reporting of S. Weltevreden from the environment, feed, animals, food, and humans in Southeast Asia is an example of how the country databank can be used to track Salmonella serotypes along the food chain.
Many serotypes are restricted to a single region of the world. This finding may reflect an ecologic niche or a local food source that is not exported. A number of such examples have been reported in the past, such as S. Marina associated with marine iguanas from South America found in the United States and S. Tilene in imported African pygmy hedgehogs in the United States and Canada (14–16). The country databank is uniquely placed to allow countries to observe this phenomenon. Investigators have reported infections of S. Javiana associated with exposure to wild amphibians in a confined area in the southeastern United States (17). According to the country databank, S. Javiana is only reported among the 15 most common serotypes in the United States. In 2001, the WHO Global Salm-Surv country databank helped confirm that S. Weltevreden was largely restricted to Southeast Asia. A survey of Southeast Asian laboratories showed that items most frequently associated with this serotype include seafood, water, and Asian vegetables (18). In the same region, S. Rissen has increased in both human and nonhuman sources (19). The country databank allows countries to become aware that a common serotype in their country may be rare elsewhere in the world, leading to hypothesis generation in outbreaks and studies to understand the sources of disease. Countries that report a large number of isolates to the country databank, such as North American and European countries, typically do not report rare serotypes because these would not rank in their top 15, thus limiting the ability to track rare serotypes in these countries.
Countries with fewer resources may lack complete antisera kits necessary to identify certain serotypes, which would lead to underreporting. For example, although we assume that S. Enteritidis human infections occur globally, a number of countries in Asia, Africa, and Latin America and the Caribbean did not report this serotype in their top 10 (Table 2). Lack of resources can also cause misclassification of serotypes. For example, S. Paratyphi B was reported to be among the most common serotypes in Latin America and the Caribbean. However, some countries in the region lack the capacity to differentiate between S. Paratyphi B and S. Paratyphi B tartrate var. Java.
In general, industrialized countries are more likely to regularly contribute to the country databank and to report more isolates. The results are therefore biased towards the industrialized world. However, the country databank lacks data from many Western European countries. Twenty-four European countries report human Salmonella serotype results annually to Enter-net, a European-based surveillance network for gastrointestinal infections, as compared to only 14 reporting to WHO Global Salm-Surv from 2000 to 2002. A review of recent Enter-net data confirms that S. Enteritidis is by far the most commonly isolated serotype in Europe but at a lower proportion than that reported to WHO Global Salm-Surv; 68%–71% of Enter-net Salmonella isolates were S. Enteritidis from 2000 to 2002 (Ian Fisher, pers. comm.) (20). A recent agreement between WHO Global Salm-Surv and Enter-net will lead to routine electronic sharing of data between the 2 systems to improve efficiency and representativeness (Henrik Wegener, pers. comm.).
Serotypes reported by a region are not necessarily circulating locally and may have been imported through travel or traded foods. Intraregional comparisons are limited by the fact that case definitions and surveillance systems vary between countries. The country databank does not collect the source of isolation. Some countries may report salmonellae isolated from both blood and stool and others from stool only. The low number of isolates and countries reporting nonhuman data and the pooling of food, animal, environmental, and feed sources hamper further analysis of nonhuman data. Some regional results may not be representative, since some regions have few countries reporting data to the country databank (e.g., Africa).
Some countries may not submit data to the country databank because of concern regarding international trading of food. Others do not have the supplies or training necessary to conduct serotyping. WHO Global Salm-Surv training courses were launched in Southeast Asia in 1999 and expanded to South America and the Middle East in 2000; China, Central America, and the Caribbean in 2001; and West Africa and Eastern Europe in 2002 (http://www.who.int/salmsurv). All participating countries were initially provided with antisera to conduct serotyping. WHO Global Salm-Surv established an external quality assurance system (EQAS) in 2000 to assess the accuracy of serotyping and antimicrobial susceptibility testing among member national reference laboratories. From 2000 to 2002, the number of laboratories participating in EQAS increased from 44 to 117, and the capacity to correctly serotype 8 Salmonella isolates improved from 76% to 90% of participating laboratories (21). The increased reporting of certain serotypes during the study period may be due to capacity improvement and increased participation in the country databank as well as real changes in the epidemiologic features of salmonellae. For example, S. Montevideo was first reported in Africa in 2001. In 2002, Tunisia participated in the country databank for the first time and reported S. Montevideo in its top 15 (increased participation). Oceania started reporting S. Montevideo in its top 15 in 2002 for the first time since participating in the country databank (change in epidemiologic features).
Data are not entered into the country databank in a timely enough way to detect international outbreaks. However, regularly monitoring the data can allow emerging trends in regional and international Salmonella epidemiology and region-specific serotypes to be detected. This information in turn leads to hypothesis generation, studies, and international collaboration to improve control of salmonellae in the long term. Examples of such work include the surveillance of Salmonella serotypes and antimicrobial drug resistance in South America, China, and the Democratic Republic of Congo; the assessment of risk factors for and drug resistance of S. Weltevreden in Southeast Asia; and the molecular characterization of S. Corvallis isolates from Bulgaria, Thailand, and Denmark (18,22–26, F. Aarestrup, pers. comm.).
The WHO Global Salm-Surv Country Databank is a valuable resource for international Salmonella surveillance. Past attempts to characterize Salmonella serotype distribution globally have either not been widely accessible or relied on irregular surveys of laboratories (8,11). Trends in global Salmonella epidemiology can now be updated and followed across regions and over time. In an era when most national institutions have access to the Internet, using a Web-based data collection tool is both feasible and practical. The data are immediately and publicly accessible for viewing and analysis (http://www.who.int/salmsurv). The results have several limitations in terms of representativeness and comparability but can be used to follow trends, generate hypotheses, and assess the effect of major interventions. This surveillance is a step toward improving the understanding and control of salmonellae worldwide.
Dr Galanis is a physician epidemiologist at the British Columbia Centre for Disease Control, Vancouver, British Columbia, Canada. Her interests and experience lie in communicable disease surveillance, prevention and control, and international health.
Acknowledgment
We thank the national reference laboratories that submitted data to the WHO Global Salm-Surv country databank, the WHO Global Salm-Surv steering committee and members, Arne Bent Jensen, and Rene Sjogren Henriksen.
References
- World Health Organization. The role of food safety in health and development: report of a joint FAO/WHO expert committee on food safety. WHO Technical Report Series, no. 705. Geneva: The Organization; 1984.
- Gomez TM, Motarjemi Y, Miyagawa S, Kaferstein FK, Stohr K. Foodborne salmonellosis. World Health Stat Q. 1997;50:81–9.PubMedGoogle Scholar
- Kaferstein FK, Motarjemi Y, Bettcher DW. Foodborne disease control: a transnational challenge. Emerg Infect Dis. 1997;3:503–10. DOIPubMedGoogle Scholar
- Todd ECD. Epidemiology of foodborne diseases: a worldwide review. World Health Stat Q. 1997;50:30–50.PubMedGoogle Scholar
- Voetsch AC, Van Gilder TJ, Angulo F, Farley MM, Shallow S, Marcus R, Foodnet estimate of the burden of illness caused by nontyphoidal Salmonella infections in the United States. Clin Infect Dis. 2004;38:S127–34. DOIPubMedGoogle Scholar
- Hald T, Vose D, Wegener HC, Koupeev T. A Bayesian approach to quantify the contribution of animal-food sources to human salmonellosis. Risk Anal. 2004;24:255–69. DOIPubMedGoogle Scholar
- Motarjemi Y, Kaferstein FK. Global estimation of foodborne diseases. World Health Stat Q. 1997;50:5–11.PubMedGoogle Scholar
- Herikstad H, Motarjemi Y, Tauxe RV. Salmonella surveillance: a global survey of public health serotyping. Epidemiol Infect. 2002;129:1–8. DOIPubMedGoogle Scholar
- Global Salm-Surv WHO. Progress report (2000–2005): building capacity for laboratory-based foodborne disease surveillance [monograph on the Internet]. [cited 2005 Dec 28]. Available from http://www.who.int/salmsurv/GSSProgressReport2005_2.pdf
- Petersen A, Aarestrup FM, Angulo FJ, Wong S, Stohr K, Wegener HC. WHO global Salm-Surv external quality assurance system (EQAS): an important step toward improving the quality of Salmonella serotyping and antimicrobial susceptibility testing worldwide. Microb Drug Resist. 2002;8:345–53. DOIPubMedGoogle Scholar
- Rodrigue DC, Tauxe RV, Rowe B. International increase in Salmonella Enteritidis: a new pandemic? Epidemiol Infect. 1990;105:21–7. DOIPubMedGoogle Scholar
- Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV. CDC PulseNet Task Force. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg Infect Dis. 2001;7:382–9.PubMedGoogle Scholar
- Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82:346–53.PubMedGoogle Scholar
- Lipsky S, Tanino T. African pygmy hedgehog-associated salmonellosis—Washington, 1994. MMWR Morb Mortal Wkly Rep. 1995;44:462–3.PubMedGoogle Scholar
- Woodward DL, Khakhria R, Johnson WM. Human salmonellosis associated with exotic pets. J Clin Microbiol. 1997;35:2786–90.PubMedGoogle Scholar
- Mermin J, Hoar B, Angulo FJ. Iguanas and Salmonella Marina infection in children: a reflection of the increasing incidence of reptile-associated salmonellosis in the United States. Pediatrics. 1997;99:399–402. DOIPubMedGoogle Scholar
- Srikantiah P, Lay JC, Crump JA, Campbell J, Van Duyne MS, Bishop R, Salmonella enterica serotype Javiana infections associated with amphibian contact, Mississippi, 2001. Epidemiol Infect. 2004;132:273–81. DOIPubMedGoogle Scholar
- Patrick ME, Hendriksen RS, Lertworapreecha M, Aarestrup FM, Chalermchaikit T, Wegener HC, Epidemiology of Salmonella Weltevreden in Southeast Asia and the Western Pacific—a WHO Global Salm-Surv regional research project [abstract]. Third International Conference on Emerging Infectious Diseases; 2004 Feb 29–Mar 3; Atlanta.
- Bangtrakulnonth A, Pornreongwong S, Pulsrikarn C, Sawanpanyalert P, Hendriksen RS, Lo Fo Wong DMA, Salmonella serovars from humans and other sources in Thailand, 1993–2002. Emerg Infect Dis. 2004;10:131–6.PubMedGoogle Scholar
- Fisher IS. International trends in Salmonella serotypes 1998–2003—a surveillance report from the Enter-net international surveillance network. Euro Surveill. 2004;9:45–7.PubMedGoogle Scholar
- Seyfarth AM, Galanis E, Hendriksen RS, Wegener HC, Jensen AB, Lo Fo Wong D, Assessing the quality of Salmonella serotyping and antimicrobial susceptibility testing in national laboratories worldwide—experiences from four years of WHO Global Salm-Surv EQAS [abstract]. Third International Conference on Emerging Infectious Diseases; 2004 Feb 29–Mar 3; Atlanta.
- WHO Global Salm-Surv South America Working Group and WHO Global Salm-Surv. A WHO Global Salm-Surv retrospective study examining serotypes in South America, 2000: dominance of Salmonella serotype Enteritidis [abstract]. Second International Conference on Emerging Infectious Diseases; 2002 Mar 24–27; Atlanta.
- WHO Global Salm-Surv South American Working Group. WHO Global Salm-Surv. WHO Global Salm-Surv (WHO-GSS) in South America, 2000–02: surveillance of Salmonella serovars and antibiotic resistance [abstract]. Third International Conference on Emerging Infectious Diseases; 2004 Feb 29–Mar 3; Atlanta.
- Wang MQ, Ran L, Xu J, Li YH, Wu SY, Yao JH, WHO Global Salm-Surv: national active surveillance for Salmonella in China: 2000–2002 [abstract]. Third International Conference on Emerging Infectious Diseases; 2004 Feb 29–Mar 3; Atlanta.
- Zono N, Vanderberg O, Mitangala P, Donnen P, Wouafo M, Aidara-Kane A, Antimicrobial susceptibility of bacterial enteric pathogens isolated in humans from 2002 to 2004 in the province of South-Kivu, Democratic Republic of Congo [abstract]. 15th European Congress of Clinical Microbiology and Infectious Diseases; 2005 Apr 2–5; Copenhagen, Denmark. Clin Microbiol Infect. 2005;11(Suppl 2):585.PubMedGoogle Scholar
- Aarestrup FM, Lertworapreecha M, Evans MC, Bangtrakulnonth A, Chalermchaikit T, Hendriksen RS, Antimicrobial susceptibility and occurrence of resistance genes among Salmonella enterica serovar Weltevreden from different countries. J Antimicrob Chemother. 2003;52:715–8. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 12, Number 3—March 2006
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Eleni Galanis, Epidemiology Services, British Columbia Centre for Disease Control, 655 West 12th Ave, Vancouver, British Columbia, V5Z 4R4, Canada; fax: 604-660-0197
Top