Volume 14, Number 10—October 2008
Perspective
Rise and Persistence of Global M1T1 Clone of Streptococcus pyogenes
Features of the Newly Emerged Hypervirulent Global M1T1 Strain
Contribution of Prophages to Emergence of Global M1T1 Strain
Acquisition of Novel Virulence Genes by Global M1T1 Strain and Effect on Virulence
In vivo Selection of Hypervirulent Descendents of Global M1T1 Strain
M1T1 and the Future of GAS Epidemiology
Cite This Article
Abstract
The resurgence of severe invasive group A streptococcal infections in the 1980s is a typical example of the reemergence of an infectious disease. We found that this resurgence is a consequence of the diversification of particular strains of the bacteria. Among these strains is a highly virulent subclone of serotype M1T1 that has exhibited unusual epidemiologic features and virulence, unlike all other streptococcal strains. This clonal strain, commonly isolated from both noninvasive and invasive infection cases, is most frequently associated with severe invasive diseases. Because of its unusual prevalence, global spread, and increased virulence, we investigated the unique features that likely confer its unusual properties. In doing so, we found that the increased virulence of this clonal strain can be attributed to its diversification through phage mobilization and its ability to sense and adapt to different host environments; accordingly, the fittest members of this diverse bacterial community are selected to survive and invade host tissue.
Group A streptococci (GAS or Streptococcus pyogenes) are strictly human pathogens that normally colonize the throat or skin without causing disease. Members of this species are differentiated into >100 types on the basis of immunogenic differences in their surface M proteins and polymorphisms in the emm gene (1). The range of GAS diseases is broad and includes both localized and systemic infections that can cause acute or chronic illnesses (Table 1 in Technical Appendix). In most cases, these bacteria cause pharyngitis (sore throat), tonsillitis, or skin infections such as impetigo/pyoderma. At times, however, the bacteria gain access to normally sterile sites and cause invasive disease. Depending on complex host–pathogen interactions, invasive GAS infections can cause either severe shock and multiple organ failure or nonsevere systemic disease, e.g., mild bacteremia and cellulitis (2,3). Likewise, invasive infections of soft tissues can be severe, e.g., necrotizing fasciitis (NF), or nonsevere, e.g., cellulitis or erysipelas (4). Whereas host genetic susceptibility plays a key role in modulating disease manifestation, variations in bacterial virulence properties contribute to infection severity.
Despite reports that particular serotypes or emm types are more commonly associated than others with particular disease manifestations, serotypic designation does not always reflect the pathogenic potential of a given strain. As we discuss below, serotype diversification can convert relatively avirulent serotypes to highly virulent ones. Dissection of molecular and genetic events leading to such diversification provides insight into how the changes in pathogenesis and host–pathogen interactions can lead to the resurgence of a severe infectious disease.
In the 19th century, GAS infections were associated with severe and frequent epidemics of invasive and often fatal illnesses, including a pandemic of scarlet fever in the United States and Great Britain (5). Invasive GAS infections with severe manifestations continued through the 1920s (5). The severity of these illnesses then declined notably until the early 1980s, when a statistically significant simultaneous recrudescence of the severe and fatal forms of invasive GAS infections occurred in different parts of the industrialized world (6,7). Accordingly, in 1993 a working group developed the case definition for streptococcal toxic-shock syndrome (STSS) as hypotension accompanied by multiple organ failure, indicated by 2 of the following signs: renal impairment, coagulopathy, liver involvement, adult respiratory distress syndrome, a generalized rash, and soft tissue necrosis (8). A modified definition of STSS was later adopted to focus on the host immune-mediated severe systemic disease associated with invasive infections, manifested by hypotension and multiple organ failure, excluding skin rash, soft-tissue necrosis, and gangrene (2). Similarly, NF was defined by the histopathologic identification of necrosis of superficial fascia and a polymorphonuclear infiltrate and edema of the reticular dermis, subcutaneous fat, and superficial fascia (9,10). The speed and rigor by which invasive GAS infections spread in the host, sometimes causing severe damage to the fascia and muscles, prompted its designation as the “flesh-eating disease.”
Epidemiologic studies showed that the resurgence of severe invasive GAS infection was not limited to sporadic cases; rather, it represented a global spread, ushering in a new pandemic, similar to that reported in the earlier part of the 20th century. An important feature of this latest pandemic is its association with a distinct epidemiologic shift in GAS serotypes. Although many GAS serotypes are capable of causing severe diseases, a few were more frequently isolated from patients with severe cases, e.g., M1, M3, M18, and M28 strains (Table 2 in Technical Appendix). However, whether those serotypes cause more severe disease because of their hypervirulence or because they were also the most prevalently isolated strains in the community at that time was not clear (11,12). These possibilities are not mutually exclusive, but in fact may be related. We believe that the unique features of the newly emerged subclones of GAS serotypes, in particular the M1T1 clonal strain, evolved as a result of diversification of the bacteria and acquisition of new genes that improved their fitness to infect humans. This, together with host-imposed pressure, resulted in the selection of hypervirulent mutants of this strain associated with an ability to cause severe forms of the invasive infection in susceptible persons.
Whereas most GAS serotypes traditionally exhibit cyclic epidemiologic patterns, appearing and disappearing from the community at different times (13), the M1T1 subclone has persisted globally for more than a quarter of a century as the most frequently isolated serotype from patients with invasive and noninvasive cases. Advanced molecular and genomic tools showed a great deal of diversity among GAS strains belonging to the same serotype, and the M1 serotype is no exception. The clonality of the reemerged M1T1 strain was first described by Cleary et al. (14) and later confirmed by others by the use of different molecular methods (Table 3 in Technical Appendix), which confirmed that the M1T1 clone differs from its ancestral M1 clone in several aspects. We will present evidence that those differences have indeed contributed to the stark difference in epidemiologic and virulence properties between 2 strains belonging to the same serotype.
Together with the Ontario Streptococcal Study Group and the Centers for Disease Control and Prevention, our laboratory launched one of the earliest and most comprehensive prospective studies of invasive GAS pathogenesis in Ontario (8), where active surveillance of invasive GAS cases took place during 1992–2002 (4,9). M1T1 isolates recovered from patients with noninvasive as well as invasive cases, of varying severity, were extensively analyzed at the molecular level and shown to be clonal regardless of case severity (3). This clonal M1T1 strain possesses the emm1.0 allele of the M1 gene (3) and is one of the opacity factor–negative GAS serotypes. This strain differs in its virulence and genomic content from other less virulent M1 strains, represented by strain M1 SF370, the first fully sequenced GAS strain (15).
Several events appear to have contributed to the diversification of the M1 GAS serotype, leading to the emergence of the M1T1 global strain. Specifically, diversification through the loss and/or acquisition of phages that took away certain genes and introduced new ones into the M1 serotype is a major contributor to the emergence of this strain. This phenomenon is certainly not unique to the M1T1 strain, but is also seen in the M3T3 and M18 strains (16,17), which co-emerged with the M1T1 clonal strain in the 1980s.
In 1996, Cleary et al. found that the globally disseminated M1T1 differs from its closely related M1 subtypes by 70 kb of phage DNA (18). Ensuing studies from our group, in which we conducted global genomic comparison of the M1T1 clones and the closely related M1 SF370 strain, demonstrated that most of the genetic differences (≈5% divergence) were accounted for by phage or phagelike sequences. After assembling these distinct sequences, we identified 2 novel prophages that were introduced into the M1T1 global strain (19). One prophage (SPhinX) carries the speA2 gene, which encodes the potent superantigen SpeA; the other (PhiRamid) carries the sda1 gene, which encodes the most potent streptococcal nuclease identified thus far (19,20). The introduction of these phages into the M1T1 clonal strain was later confirmed by the complete genome sequence of a clinical M1T1 isolate, MGAS5005 (21).
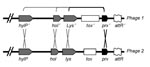
The M1T1 prophages exhibit considerable genetic mosaicism, and the sequence analysis of the 2 novel M1T1 phages demonstrates that these bacterial viruses continuously exchange functional modules by various genetic mechanisms, including different modes of recombination (19). We believe that exchange between the lysis and lysogenic conversion modules of GAS prophages has led to the swapping of virulence genes (toxins) among phages (19). We also believe that this process is facilitated by a highly conserved gene, paratox (prx), commonly found between the toxin gene and phage attachment site. Conserved prx sequences on 1 side of the toxin gene together with 1–3 highly conserved phage genes on the other side (lysin, holin, and/or hyaluronidase genes) are likely to facilitate recombination events leading to swapping of toxin genes among bacterial isolates (Figure 1) (19). This notion is supported by the fact that strains belonging to the same serotype may have different virulence components carried by the same or highly similar phages, whereas those belonging to different serotypes may, in fact, have identical phage-encoded toxins. For example, 4 highly similar phages (370.3, 5005.2, MemPhiS, 315.3) identified in M1 SF370, M1T1 5005, M1T1 6050, and M3 strains, respectively, have different DNases in their lysogenic conversion modules. Phages 370.3 and 5005.2 are >99% identical to each other and carry the mf3 gene, and each is 90% identical to MemPhiS and 315.3, which carry the mf4 gene instead (Figure 2).
Two virulence genes, speA2 and sda1, were introduced into the M1T1 strain by prophages and are likely to have contributed to its increased fitness and virulence (19,21). SpeA2 is an important and potent streptococcal superantigen. Although GAS has a rich superantigen repertoire, different strains harbor different combinations of superantigen genes—some are phage encoded, while others are integrated into the bacterial chromosome. Both the global M1T1 strain and its ancestral SF370 (15) strain have the superantigen-encoding genes speF, speG, speJ, and smeZ1. However, these 2 strains differ in that the global M1T1 strain has speA2, whereas SF370 has speC (Table). Both SpeA and SpeC are prophage-encoded, whereas the other M1T1 superantigens are chromosomal. Additionally, the clonal M1T1 strain lacks speH and speI, which are encoded on a single phage in M1 SF370 (15,21).
Of particular relevance to this discussion is that the speA gene was seen in M1 isolates obtained in the early 20th century but had almost vanished from M1 isolates obtained between the 1920s and early 1980s. The loss of speA was thought to be one of the main reasons for the sharp decline in severe invasive GAS infections during this time (23,24). Likewise, the reintroduction of the speA2 allele in the M1T1 clonal strain in the 1980s prompted speculations that SpeA, and in particular its allelic variant SpeA2, was a major factor in the resurgence of severe invasive GAS infections during that time (25). However, additional studies showed that, although the reintroduction of speA may have been a factor, the acquisition of other virulence genes by the M1T1 clone is more likely to have had a more profound effect on its increased fitness and virulence in vivo (20,26,27). Nonetheless, the fact that SpeA was missing from most GAS isolates for >50 years suggests that the reintroduction of this superantigen may have increased the risk for persons to have invasive infections because they lack antibodies that neutralize its superantigenic activity. Indeed, the lack of superantigen-neutralizing antibodies has been shown to increase the risk for invasive disease (28,29).
Sda1, which was also acquired by the M1T1 global strain, is a potent streptodornase (streptococcal nuclease) (20) and is not found in most of the other prevalent strains but has been recently reported in an M12 strain (16). Streptodornases are secreted extracellular nucleases classically thought to play an important role in virulence by degrading pus (30). Every GAS serotype sequenced so far contains >1 streptodornase paralog. These various streptodornases, which differ in the pH optima for their nuclease activity, are likely functionally nonredundant, possibly having different substrate specificity, and may be differentially active in certain host niches or at different times during the infection. The M1T1 clone has, in addition to sda1, the chromosomal streptodornase spd/mf (alias speF) and another phage-encoded streptodornase, spd3/mf3 (or—less frequently—spd4/mf4); however, it lacks spd2/mf2 found in the M1 SF370 (19,20). Despite the presence of multiple DNases in the bacteria, Sda1 has the highest specific activity among the streptococcal nucleases. We showed that the increased activity of Sda1 has resulted from a frame-shift mutation in its C terminus, and when the additional C-terminal sequence of Sdal was deleted, the enzyme activity dropped significantly (20).
Sda1, unlike the other nucleases, appears to play a major role in virulence, and inactivating its gene resulted in a dramatic loss of virulence (26,27,31), whereas introducing it into an avirulent strain led to a virulent phenotype (26). Sdal protects bacteria against neutrophils (31)—which entrap the bacteria in neutrophil extracellular traps (NETs) (32)—by degrading these DNA NETs, thereby freeing the bacteria and promoting their ability to invade host tissues (26). Additionally, recent evidence suggests that the Sda1 expression may synergize with host factors, leading to additional selective pressure on the bacteria in vivo and resulting in the emergence of a hypervirulent phenotype of the same bacteria (27).
Besides the exchange of phage-encoded toxins, additional recombination events may have contributed to the diversification of the M1T1 clone. In a recent study, Sumby et al. (21) used DNA–DNA hybridization and single nucleotide polymorphism analysis to show that a 36-kb chromosomal region has been horizontally transferred to M1T1 by recombinatorial replacement from an M12 ancestral strain. This chromosomal region harbors genes encoding 2 important toxins, streptolysin O (SLO) and nicotinamide glycohydrolase (NADGH or NADase), both of which were more highly expressed in M1T1-MGAS5005 compared to M1 SF370 (21). Although these differences in expression might be a consequence of the recombination event, we believe that the enhanced expression of these genes is more likely due to a mutation in the covS gene of the studied MGAS5005 strain, which resulted in higher expression of virulence networks (33). SLO is an important GAS cytolysin that enhances cytotoxicity and toxin translocation (34,35), and its heightened expression would be expected to increase virulence. It is therefore apparent that several mechanisms led to GAS diversification and that the globally disseminated M1T1 clone has acquired several virulence factors that seem to have contributed to its unusual persistence, spread, and virulence.
In addition to the introduction and loss of specific genes in the global M1T1 strain, a high degree of variability in the expression of virulence genes among isolates belonging to this clonal strain was reported (3,36). This variable expression, in part, depended on where and when the isolates were recovered from the host. However, one of the most notable changes in gene expression that arises in response to host environmental pressure is the remarkable downregulation of the major streptococcal protease, SpeB, and the consequent significant increase in bacterial invasion and severity of GAS sepsis (27,37,38).
Earlier studies by Kansal et al. (37) provided the first hint for the reciprocal relation between SpeB expression and severity of GAS sepsis, when they observed that isolates recovered from patients with more severe cases expressed no, or significantly less, SpeB compared to those recovered from patients with nonsevere cases. In ensuing studies, we found that M1T1 regulates its secreted proteins by at least 2 mechanisms (39), a transcriptional regulation, and a posttranslational degradation and remodeling of bacterial proteins by SpeB, that, itself, is tightly regulated ( 40; Technical Appendix, supplementary reference 41). The secreted proteome in the presence and absence of active SpeB is starkly different. Essentially most extracellular virulence factors, including M protein, streptokinase, SpeF, Sda1, C5a peptidase, and the secreted inhibitor of complement, are degraded by this protease, resulting in decreased virulence. The advantage of this massive degradation of virulence factors to the bacteria is not entirely known, but we predict that this may be a means by which the bacteria camouflage themselves from the host during the initial stages of infection. By degrading their virulence components, bacteria may evade initial innate host defenses at the site of the infection until they gain access to a host niche (e.g., skin), where they can start to multiply. Thus, SpeB may facilitate the initial invasion of bacteria through its proteolytic action on host matrix proteins. However, within 60–80 hours after infection, the bacteria are subjected to a hostile human environment, and consequently, there is a selection for more fit mutants within the bacterial community that are better adapted to confront host defenses and gain access to blood and possibly other sterile sites. The more fit mutants, it turns out, are those that lack SpeB expression because of a mutation in covS, which is a part of a 2-component regulatory system (CovRS) involved in regulating 15% of GAS genes including SpeB (Technical Appendix, supplementary reference 42). Indeed, recent studies provided evidence for the co-existence of at least 2 very different phenotypic forms of M1T1 bacteria in the initial stages of infection through the skin of mice characterized by SpeB+ or SpeB– phenotypes (27,39).
The downregulation of SpeB spares several key virulence factors that include Sda1 and streptokinase. Our recent studies showed that sparing Sda1 frees the bacteria from neutrophil NETs (27). Similarly, in a human plasminogen-transgenic mouse model, sparing streptokinase allowed accumulation of surface plasmin activity and increased bacterial evasion (Technical Appendix, supplementary reference 43). Additional differentially expressed genes in the in vivo–selected covS mutants are also likely to contribute to increased virulence, and these are currently being investigated.
Is there an exit plan for M1T1? How long will this strain survive and prevail? Will there be another prevalent strain in the future? It is intriguing that although M1T1 causes deadly conditions, this clone keeps infecting many persons, retaining its superior prevalence. This suggests that there is an exit plan for this clone, or that it is so widely spread among the human population that it keeps being transmitted through genetically protected persons, who serve as reservoirs for it. The diversity within the bacterial population in the host also suggests that while hypervirulent mutants cause deadly diseases when the bacteria invade unusual niches, the less virulent members of the same population survive well in the primary niche (e.g., the throat or nasopharynx) and thus could drive the disease transmission.
Several potentially interactive factors may have contributed to the persistence of M1T1 and may maintain this strain for a long time. These factors include the acquisition of new virulence genes and the differential regulation and expression of virulence genes caused by selection of mutants within the microbial community. These changes in the pathogen, as well as changes in herd immunity and differential host susceptibility, are likely to create dynamic interactions between streptococci and their human host.
When novel strains or clones emerge that express novel proteins or variants of old proteins, these strains are endowed with the ability to better withstand the pressure of herd immunity. According to this hypothesis, M1T1 and other strains that reemerged in the mid-1980s may have successfully survived herd immunity either because they acquired new protein-encoding genes or because they possessed allelic variants of key genes encoding proteins and/or novel alleles that were as-yet unsampled by the immune system. Also, the acquisition of new genes or the sparing of existing proteins from proteolytic degradation may have endowed the bacteria with means to better evade host immune defenses.
In summary, we believe that the emergence of the M1T1 strain, its diversification by phage acquisition, and the in vivo selection of more fit members of its community present an intriguing example of molecular events that can drastically change the epidemiology and virulence of an otherwise avirulent or less virulent organism. Predicting whether other GAS strains may follow a similar trajectory to M1T1 is difficult: The next prevalent strain to emerge may have to combine changes in chromosomal and phage-encoded genes to enhance its fitness and allow it to adapt to different host environments; it also has to be resistant enough to phage-driven lysis. (More prophages enrich the bacteria with additional toxins, but they may also bring the potential risk of lysing the bacteria at any time a phage is induced.) As there are now more and more examples of phage exchange even within and between different bacterial species (Technical Appendix, supplementary reference 44), the traditional classification schema may have to be replaced by ones that better reflect the bacterial virulome. This virulome, as discussed here, can be grossly altered, depending on the environment the bacteria face and the consequent selection of underrepresented minority of the bacterial community that is best adapted to deal with various hostile host milieus.
Dr Aziz received his PhD in microbiology and immunology in 2005 from the University of Tennessee Health Science Center, Memphis. He is currently a lecturer in the Department of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Cairo, Egypt, and an adjunct faculty member at San Diego State University, San Diego, California.
Dr Kotb is the chair of the Department of Molecular Genetics, Biochemistry, and Microbiology at the University of Cincinnati, Cincinnati, Ohio. She is also a joint professor in the Department of Molecular Sciences and has directed the Mid-South Center for Biodefense and Security at the University of Tennessee Health Science Center in Memphis. Dr. Kotb has led a team of investigators who, since 1990, have been studying the reemergence of highly severe forms of invasive GAS infections.
References
- Facklam RF, Martin DR, Lovgren M, Johnson DR, Efstratiou A, Thompson TA, Extension of the Lancefield classification for group A streptococci by addition of 22 new M protein gene sequence types from clinical isolates: emm103 to emm124. Clin Infect Dis. 2002;34:28–38. DOIPubMedGoogle Scholar
- Kotb M, Norrby-Teglund A, McGeer A, El-Sherbini H, Dorak MT, Khurshid A, An immunogenetic and molecular basis for differences in outcomes of invasive group A streptococcal infections. Nat Med. 2002;8:1398–404. DOIPubMedGoogle Scholar
- Chatellier S, Ihendyane N, Kansal RG, Khambaty F, Basma H, Norrby-Teglund A, Genetic relatedness and superantigen expression in group A Streptococcus serotype M1 isolates from patients with severe and nonsevere invasive diseases. Infect Immun. 2000;68:3523–34. DOIPubMedGoogle Scholar
- Davies HD, McGeer A, Schwartz B, Green K, Cann D, Simor AE, Invasive group A streptococcal infections in Ontario, Canada. N Engl J Med. 1996;335:547–54. DOIPubMedGoogle Scholar
- Katz AR, Morens DM. Severe streptococcal infections in historical perspective. Clin Infect Dis. 1992;14:298–307.PubMedGoogle Scholar
- Hoge CW, Schwartz B, Talkington DF, Breiman RF, MacNeill EM, Englender SJ. The changing epidemiology of invasive group A streptococcal infections and the emergence of streptococcal toxic shock-like syndrome. A retrospective population-based study. JAMA. 1993;269:384–9. DOIPubMedGoogle Scholar
- Low DE, Schwartz B, McGeer A. The reemergence of severe group A streptococcal disease: an evolutionary perspective. In: Scheld WM, Armstrong D, Hughes JM, editors. Emerging infections. Washington: ASM Press; 1997. p. 93–123.
- Infections WGoSS. Defining the group A streptococcal toxic shock syndrome. Rationale and consensus definition. The Working Group on Severe Streptococcal Infections. JAMA. 1993;269:390–1. DOIPubMedGoogle Scholar
- Kaul R, McGeer A, Low DE, Green K, Schwartz B. Population-based surveillance for group A streptococcal necrotizing fasciitis: clinical features, prognostic indicators, and microbiologic analysis of seventy-seven cases. Ontario Group A Streptococcal Study. Am J Med. 1997;103:18–24. DOIPubMedGoogle Scholar
- Sharkawy A, Low DE, Saginur R, Gregson D, Schwartz B, Jessamine P, Severe group A streptococcal soft-tissue infections in Ontario: 1992–1996. Clin Infect Dis. 2002;34:454–60. DOIPubMedGoogle Scholar
- Johnson DR, Wotton JT, Shet A, Kaplan EL. A comparison of group A streptococci from invasive and uncomplicated infections: Are virulent clones responsible for serious streptococcal infections? J Infect Dis. 2002;185:1586–95. DOIPubMedGoogle Scholar
- Rogers S, Commons R, Danchin MH, Selvaraj G, Kelpie L, Curtis N, Strain prevalence, rather than innate virulence potential, is the major factor responsible for an increase in serious group A Streptococcus infections. J Infect Dis. 2007;195:1625–33. DOIPubMedGoogle Scholar
- Ikebe T, Murai N, Endo M, Okuno R, Murayama S, Saitoh K, Changing prevalent T serotypes and emm genotypes of Streptococcus pyogenes isolates from streptococcal toxic shock-like syndrome (TSLS) patients in Japan. Epidemiol Infect. 2003;130:569–72.PubMedGoogle Scholar
- Cleary PP, Kaplan EL, Handley JP, Wlazlo A, Kim MH, Hauser AR, Clonal basis for resurgence of serious Streptococcus pyogenes disease in the 1980s. Lancet. 1992;339:518–21. DOIPubMedGoogle Scholar
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci U S A. 2001;98:4658–63. DOIPubMedGoogle Scholar
- Beres SB, Musser JM. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One. 2007;2:e800. DOIPubMedGoogle Scholar
- Beres SB, Sylva GL, Sturdevant DE, Granville CN, Liu M, Ricklefs SM, Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infections. Proc Natl Acad Sci U S A. 2004;101:11833–8. DOIPubMedGoogle Scholar
- Cleary PP, LaPenta D, Vessela R, Lam H, Cue D. A globally disseminated M1 subclone of group A streptococci differs from other subclones by 70 kilobases of prophage DNA and capacity for high-frequency intracellular invasion. Infect Immun. 1998;66:5592–7.PubMedGoogle Scholar
- Aziz RK, Edwards RA, Taylor WW, Low DE, McGeer A, Kotb M. Mosaic prophages with horizontally acquired genes account for the emergence and diversification of the globally disseminated M1T1 clone of Streptococcus pyogenes. J Bacteriol. 2005;187:3311–8. DOIPubMedGoogle Scholar
- Aziz RK, Ismail SA, Park HW, Kotb M. Post-proteomic identification of a novel phage-encoded streptodornase, Sda1, in invasive M1T1 Streptococcus pyogenes. Mol Microbiol. 2004;54:184–97. DOIPubMedGoogle Scholar
- Sumby P, Porcella SF, Madrigal AG, Barbian KD, Virtaneva K, Ricklefs SM, Evolutionary origin and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–82. DOIPubMedGoogle Scholar
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–702. DOIPubMedGoogle Scholar
- Cone LA, Woodard DR, Schlievert PM, Tomory GS. Clinical and bacteriologic observations of a toxic shock-like syndrome due to Streptococcus pyogenes. N Engl J Med. 1987;317:146–9.PubMedGoogle Scholar
- Schlievert PM, Bettin KM, Watson DW. Production of pyrogenic exotoxin by groups of streptococci: association with group A. J Infect Dis. 1979;140:676–81.PubMedGoogle Scholar
- Nelson K, Schlievert PM, Selander RK, Musser JM. Characterization and clonal distribution of four alleles of the speA gene encoding pyrogenic exotoxin A (scarlet fever toxin) in Streptococcus pyogenes. J Exp Med. 1991;174:1271–4. DOIPubMedGoogle Scholar
- Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr Biol. 2006;16:396–400. DOIPubMedGoogle Scholar
- Walker MJ, Hollands A, Sanderson-Smith ML, Cole JN, Kirk JK, Henningham A, DNase Sda1 provides selection pressure for a switch to invasive group A streptococcal infection. Nat Med. 2007;13:981–5. DOIPubMedGoogle Scholar
- Basma H, Norrby-Teglund A, Guedez Y, McGeer A, Low DE, El-Ahmedy O, Risk factors in the pathogenesis of invasive group A streptococcal infections: role of protective humoral immunity. Infect Immun. 1999;67:1871–7.PubMedGoogle Scholar
- Norrby-Teglund A, Kaul R, Low DE, McGeer A, Newton DW, Andersson J, Plasma from patients with severe invasive group A streptococcal infections treated with normal polyspecific IgG inhibits streptococcal superantigen-induced T cell proliferation and cytokine production. J Immunol. 1996;156:3057–64.PubMedGoogle Scholar
- Tillet WS, Sherry S, Christensen LR. Streptococcal desoxyribonuclease: significance in lysis of purulent exudates and production by srains of hemolytic streptococci. Proc Soc Exp Biol Med. 1948;68:184–8.PubMedGoogle Scholar
- Sumby P, Barbian KD, Gardner DJ, Whitney AR, Welty DM, Long RD, Extracellular deoxyribonuclease made by group A Streptococcus assists pathogenesis by enhancing evasion of the innate immune response. Proc Natl Acad Sci U S A. 2005;102:1679–84. DOIPubMedGoogle Scholar
- Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–5. DOIPubMedGoogle Scholar
- Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group A streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. DOIPubMedGoogle Scholar
- Savic DJ, McShan WM, Ferretti JJ. Autonomous expression of the slo gene of the bicistronic nga-slo operon of Streptococcus pyogenes. Infect Immun. 2002;70:2730–3. DOIPubMedGoogle Scholar
- Ghosh J, Caparon MG. Specificity of Streptococcus pyogenes NAD(+) glycohydrolase in cytolysin-mediated translocation. Mol Microbiol. 2006;62:1203–14. DOIPubMedGoogle Scholar
- Talkington DF, Schwartz B, Black C, Todd J, Elliott J, Breiman R, Association of phenotypic and genotypic characteristics of invasive Streptococcus pyogenes isolates with clinical components of streptococcal toxic shock syndrome. Infect Immun. 1993;61:3369–74.PubMedGoogle Scholar
- Kansal RG, McGeer A, Low DE, Norrby-Teglund A, Kotb M. Inverse relation between disease severity and expression of the streptococcal cysteine protease, SpeB, among clonal M1T1 isolates recovered from invasive group A streptococcal infection cases. Infect Immun. 2000;68:6362–9. DOIPubMedGoogle Scholar
- Cole JN, McArthur JD, McKay FC, Sanderson-Smith ML, Cork AJ, Ranson M, Trigger for group A streptococcal M1T1 invasive disease. FASEB J. 2006;20:1745–7. DOIPubMedGoogle Scholar
- Aziz RK, Pabst MJ, Jeng A, Kansal R, Low DE, Nizet V, Invasive M1T1 group A Streptococcus undergoes a phase-shift in vivo to prevent proteolytic degradation of multiple virulence factors by SpeB. Mol Microbiol. 2004;51:123–34. DOIPubMedGoogle Scholar
- Lyon WR, Gibson CM, Caparon MG. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–75. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 14, Number 10—October 2008
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Ramy K. Aziz, Department of Microbiology and Immunology, Faculty of Pharmacy, Cairo University, Kasr El-Aini St, 11562 Cairo, Egypt;
Top