Volume 14, Number 12—December 2008
Letter
Candidate New Species of Kobuvirus in Porcine Hosts
Figure
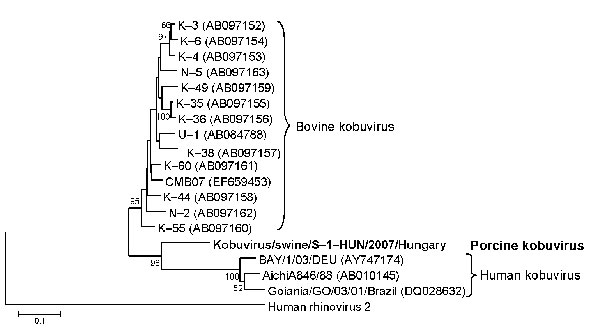
Figure. Phylogenetic tree of porcine kobuvirus (Kobuvirus/swine/S-1-HUN/2007/Hungary, GenBank accession no. EU787450), based upon the 1,065-nt fragment of the kobuvirus 3C/3D regions. The phylogenetic tree was constructed by using the neighbor-joining clustering method; distance was calculated by using the maximum composite likelihood correction for evolutionary rate with help of the MEGA version 4.0 software (10). Bootstrap values (based on 1,000 replicates) for each node if >50% are given. Reference strains were obtained from GenBank. The human rhinovirus 2 strain (X02316) was included in the tree as an outgroup. Scale bar indicates nucleotide substitutions per site.
References
- Online ICTV. Discussion forum of the International Committee on Taxonomy of Viruses, ICTV 2008 Official Taxonomy [cited 2008 Oct 22]. Available from http://talk.ictvonline.org
- Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J Infect Dis. 1991;164:954–7.PubMedGoogle Scholar
- Yamashita T, Ito M, Kabashima Y, Tsuzuki H, Fujiura A, Sakae K. Isolation and characterization of a new species of kobuvirus associated with cattle. J Gen Virol. 2003;84:3069–77. DOIPubMedGoogle Scholar
- Pham NT, Khamrin P, Nguyen TA, Kanti DS, Phan TG, Okitsu S, Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan, Bangladesh, Thailand, and Vietnam. J Clin Microbiol. 2007;45:2287–8. DOIPubMedGoogle Scholar
- Oh DY, Silva PA, Hauroeder B, Deidrich S, Cardoso DD, Schreier E. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch Virol. 2006;151:1199–206. DOIPubMedGoogle Scholar
- Ambert-Balay K, Lorrot M, Bon F, Giraudon H, Kaplon J, Wolfer M, Prevalence and genetic diversity of Aichi virus strains in stool samples from community and hospitalized patients. J Clin Microbiol. 2008;46:1252–8. DOIPubMedGoogle Scholar
- Khamrin P, Maneekarn N, Peerakome S, Okitsu S, Mizuguchi M, Ushijama H. Bovine kobuviruses from cattle with diarrhea. Emerg Infect Dis. 2008;14:985–6. DOIPubMedGoogle Scholar
- Jiang X, Huang PW, Zhong WM, Farkas T, Cubitt DW, Matson DO. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J Virol Methods. 1999;83:145–54. DOIPubMedGoogle Scholar
- Reuter G, Krisztalovics K, Vennema H, Koopmans M. Szűcs Gy. Evidence of the etiological predominance of norovirus in gastroenteritis outbreaks—emerging new variant and recombinant noroviruses in Hungary. J Med Virol. 2005;76:598–607. DOIPubMedGoogle Scholar
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
Page created: July 22, 2010
Page updated: July 22, 2010
Page reviewed: July 22, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.