Highly Pathogenic Avian Influenza Virus (H5N1) in Domestic Poultry and Relationship with Migratory Birds, South Korea
Youn-Jeong Lee*
1, Young-Ki Choi†
1, Min-Suk Song†, Ok-Mi Jeong*, Eun-Kyoung Lee*, Woo-Jin Jeon*, Wooseog Jeong*, Seong-Joon Joh*, Kang-seuk Choi*, Moon Her*, Eun ho Lee†, Tak-Gue Oh†, Ho-Jin Moon‡, Dae-Won Yoo‡, Chul-Joong Kim‡

, Moon-Hee Sung¶, Haryoung Poo#, Jun-Hun Kwon*, Jae-Hong Kim§

, Yong-Joo Kim*

, Min-Chul Kim*

, Aeran Kim*

, and Min-Jeong Kim*

Author affiliations: *Ministry of Agriculture and Forestry, Anyang, South Korea; †College of Medicine and Medical Research Institute of Chungbuk National University, Cheongju, South Korea; ‡College of Veterinary Medicine of Chungnam National University, Daejeon, South Korea; §College of Veterinary Medicine of Seoul National University, Seoul, South Korea; ¶Bioleaders Corporation, Daejeon, South Korea; #Korea Research Institute of Bioscience and Biotechnology, Daejeon, South Korea;
Main Article
Figure 2
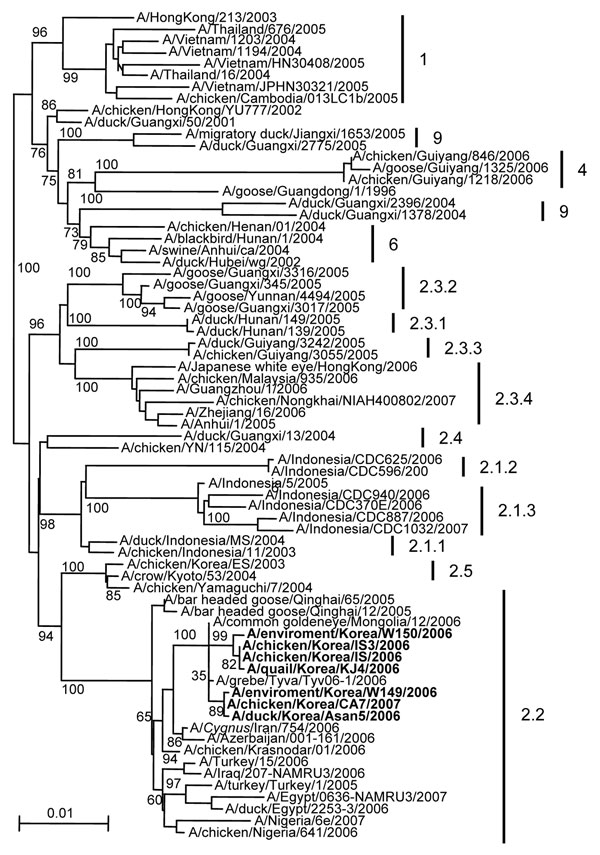
Figure 2. Phylogenetic trees for hemagglutinin (HA) genes of Korean influenza virus (H5N1) isolates from wild birds and poultry farms during 2006–2007. The DNA sequences were compiled and edited by using the Lasergene sequence analysis software package (DNASTAR, Madison, WI, USA). Multiple sequence alignments were made by using ClustalX (10). Rooted phylograms were prepared with the neighbor-joining algorithm and then plotted by using NJplot (11). Branch lengths are proportional to sequence divergence and can be measured relative to the scale bar shown (0.01-nt changes per site). Branch labels record the stability of the branches >1,000 bootstrap replicates. The tree was produced by referring to the proposed global nomenclature system for influenza virus (H5N1) (www.offlu.net). Boldface indicates isolates tested in the current study.
Main Article
Page created: July 07, 2010
Page updated: July 07, 2010
Page reviewed: July 07, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
